
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,359,621 – 2,359,730 |
| Length | 109 |
| Max. P | 0.779079 |

| Location | 2,359,621 – 2,359,730 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.44004 |
| G+C content | 0.60990 |
| Mean single sequence MFE | -38.54 |
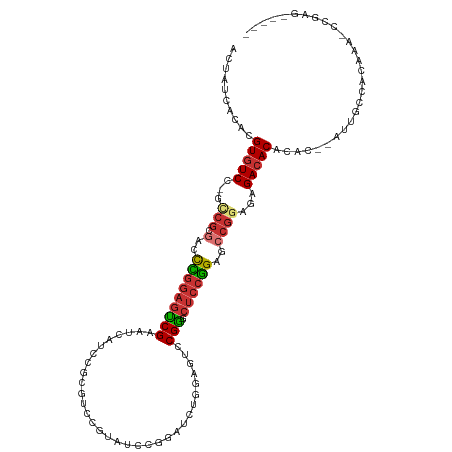
| Consensus MFE | -18.13 |
| Energy contribution | -18.96 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
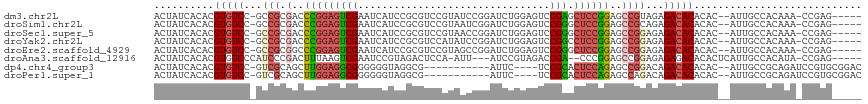
>dm3.chr2L 2359621 109 + 23011544 ACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAUCCGGAUCUGGAGUCCGAGCUCCGGAGCCGUAGAGACACACAC--AUUGCCACAAA-CCGAG----- ..........(((((.-.(..((.((((((((((.....((((.(((((....))))).))))...))).)))))).).))..).)))))....--...........-.....----- ( -36.80, z-score = -1.78, R) >droSim1.chr2L 2316330 109 + 22036055 ACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAAUCGGAUCUGGAGUCCGGGCUCCGGAGCCGCAGAGACACACAC--AUUGCCACAAA-CCGAG----- ..........((((((-((.((...((((((((......((((.(((((....))))).)))).....)))))))).)).)).).)))))....--...........-.....----- ( -39.40, z-score = -1.85, R) >droSec1.super_5 531238 109 + 5866729 ACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAACCGGAUCUGGAGUCCGGGCUCCGGAGCCGGAGAGACACACAC--AUUGCCACAAA-CCGAG----- ..........((((((-.(((.(..((((((((......((((.(((((....))))).)))).....))))))))..)))).).)))))....--...........-.....----- ( -41.00, z-score = -1.82, R) >droYak2.chr2L 2342056 109 + 22324452 ACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCAUAUCCGGAUCUGGAGUCCGGCCUCCGGAGCCGGAGAGACACACAC--AUUGCCACAAA-CCGAG----- ..........((((((-.(((.(..(((((((((.....((((.((((......)))).))))...))).))))))..)))).).)))))....--...........-.....----- ( -37.70, z-score = -1.72, R) >droEre2.scaffold_4929 2398829 109 + 26641161 ACUAUCACACGUGUCC-GCCGCGGCCCGGAGUCGAAUCAUCCGCGUCCGUAGCCGGAUCUGGAGUCCGGGCUCCGGAGCCGGAGAGACACACAC--AUUGCCACAAA-CCGAG----- ..........((((((-..(.((((((((((((......((((.(((((....))))).)))).....)))))))).))))).).)))))....--...........-.....----- ( -43.00, z-score = -1.66, R) >droAna3.scaffold_12916 10083743 106 - 16180835 ACUAUCACACGUGUCCCAUCCCGACUUUAAGUCGAAUCCGUAGACUCCA-AUU---AUCCGUAGACCGA--CCCGGAGCCGGAGAGAGACACACUCAUUGCCACAUA-CCGAG----- ..........(((((.(....((((.....))))..........((((.-...---.((((........--..))))...)))).).)))))...............-.....----- ( -21.50, z-score = -0.78, R) >dp4.chr4_group3 7656093 100 - 11692001 ACUAUCACACGUGUCC-GUCGCAGCUUGGAGGCGGGGGGUAGGCG-----------AUUC----UCCGCACUCCAGAGCCGGACAGACACACAC--AUUGCCGCAGAUCCGUGCGGAC ..........(((((.-(((.(.(((((((((((((((((.....-----------))))----))))).))))).))).)))).)))))....--....(((((......))))).. ( -44.60, z-score = -3.10, R) >droPer1.super_1 4774141 100 - 10282868 ACUAUCACACGUGUCC-GUCGCAGCUUGGAGGCGGGGGGUAGGCG-----------AUUC----UCCGCACUCCAGAGCCAGACAGACACACAC--AUUGCCGCAGAUCCGUGCGGAC ..........(((((.-(((...(((((((((((((((((.....-----------))))----))))).))))).)))..))).)))))....--....(((((......))))).. ( -44.30, z-score = -3.44, R) >consensus ACUAUCACACGUGUCC_GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAUCCGGAUCUGGAGUCCGGGCUCCGGAGCCGGAGAGACACACAC__AUUGCCACAAA_CCGAG_____ ..........(((((...(((.(..(((((((((..........((((......))))........))).))))))..))))...)))))............................ (-18.13 = -18.96 + 0.83)

| Location | 2,359,621 – 2,359,730 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.44004 |
| G+C content | 0.60990 |
| Mean single sequence MFE | -45.41 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.51 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
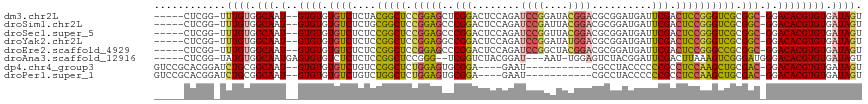
>dm3.chr2L 2359621 109 - 23011544 -----CUCGG-UUUGUGGCAAU--GUGUGUGUCUCUACGGCUCCGGAGCUCGGACUCCAGAUCCGGAUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGU -----.....-.((((.(((.(--((((.((((....((((.((((((.(((((((((.(.((((....)))))..))).).))))))))))))))).)))-).)))))))).)))). ( -46.80, z-score = -1.49, R) >droSim1.chr2L 2316330 109 - 22036055 -----CUCGG-UUUGUGGCAAU--GUGUGUGUCUCUGCGGCUCCGGAGCCCGGACUCCAGAUCCGAUUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGU -----.....-.((((.(((..--...((((((((((((((.((((((..((((((((.(.((((....)))))..))).).)))).))))))))))))).-)))))))))).)))). ( -47.00, z-score = -1.33, R) >droSec1.super_5 531238 109 - 5866729 -----CUCGG-UUUGUGGCAAU--GUGUGUGUCUCUCCGGCUCCGGAGCCCGGACUCCAGAUCCGGUUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGU -----.....-.((((.(((.(--((((.((((....((((.((((((..((((((((.(.((((....)))))..))).).)))).)))))))))).)))-).)))))))).)))). ( -44.30, z-score = -0.25, R) >droYak2.chr2L 2342056 109 - 22324452 -----CUCGG-UUUGUGGCAAU--GUGUGUGUCUCUCCGGCUCCGGAGGCCGGACUCCAGAUCCGGAUAUGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGU -----(.(((-(((((.((...--((.(((((((.(((((.((.((((......)))).)).)))))...))))))).))((((((....)))))))).))-)))).)).)....... ( -45.50, z-score = -0.79, R) >droEre2.scaffold_4929 2398829 109 - 26641161 -----CUCGG-UUUGUGGCAAU--GUGUGUGUCUCUCCGGCUCCGGAGCCCGGACUCCAGAUCCGGCUACGGACGCGGAUGAUUCGACUCCGGGCCGCGGC-GGACACGUGUGAUAGU -----.....-.((((.(((.(--((((.((((....((((.((((((..((((((((.(.((((....)))))..))).).)))).)))))))))).)))-).)))))))).)))). ( -47.00, z-score = -0.76, R) >droAna3.scaffold_12916 10083743 106 + 16180835 -----CUCGG-UAUGUGGCAAUGAGUGUGUCUCUCUCCGGCUCCGGG--UCGGUCUACGGAU---AAU-UGGAGUCUACGGAUUCGACUUAAAGUCGGGAUGGGACACGUGUGAUAGU -----.....-..(((.(((.....((((((((..(((((((..(((--((.(((....)))---...-..(((((....))))))))))..)))))))..))))))))))).))).. ( -41.80, z-score = -2.82, R) >dp4.chr4_group3 7656093 100 + 11692001 GUCCGCACGGAUCUGCGGCAAU--GUGUGUGUCUGUCCGGCUCUGGAGUGCGGA----GAAU-----------CGCCUACCCCCCGCCUCCAAGCUGCGAC-GGACACGUGUGAUAGU (.(((((......))))))...--(..(((((((((((((((.(((((.((((.----(...-----------........).)))))))))))))).)))-)))))))..)...... ( -46.90, z-score = -3.45, R) >droPer1.super_1 4774141 100 + 10282868 GUCCGCACGGAUCUGCGGCAAU--GUGUGUGUCUGUCUGGCUCUGGAGUGCGGA----GAAU-----------CGCCUACCCCCCGCCUCCAAGCUGCGAC-GGACACGUGUGAUAGU (.(((((......))))))...--(..(((((((((((((((.(((((.((((.----(...-----------........).)))))))))))))).)))-)))))))..)...... ( -44.00, z-score = -2.73, R) >consensus _____CUCGG_UUUGUGGCAAU__GUGUGUGUCUCUCCGGCUCCGGAGCCCGGACUCCAGAUCCGGAUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC_GGACACGUGUGAUAGU ........................(..(((((((...(((((.(((((..(((........((((....))))..........))).)))))))))).....)))))))..)...... (-22.74 = -22.51 + -0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:01 2011