| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,344,133 – 2,344,223 |
| Length | 90 |
| Max. P | 0.967654 |

| Location | 2,344,133 – 2,344,223 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Shannon entropy | 0.30511 |
| G+C content | 0.53066 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
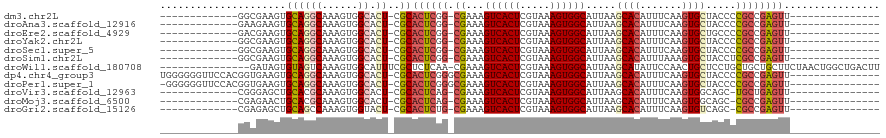
>dm3.chr2L 2344133 90 + 23011544 -------------GGCGAAGUGCAGGCAAAGUGGCACU-CGCACUCGG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU--------------- -------------.((((.((((..((...)).)))))-)))((((((-((...((((((.....))))))....(((((.......)))))....)))))))).--------------- ( -38.80, z-score = -4.36, R) >droAna3.scaffold_12916 10064900 90 - 16180835 -------------GAAGAAGUGCAGGCAAAGUGGCACU-CGCACUCGG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU--------------- -------------...((.((((..((...)).)))))-)..((((((-((...((((((.....))))))....(((((.......)))))....)))))))).--------------- ( -33.20, z-score = -3.24, R) >droEre2.scaffold_4929 2383216 90 + 26641161 -------------GACGAAGUGCAGGCAAAGUGGCACU-CGCACUCGG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUGCCCCGCCGAGUU--------------- -------------(.(((.((((..((...)).)))))-)))((((((-((...((((((.....))))))....(((((.......)))))....)))))))).--------------- ( -35.00, z-score = -2.93, R) >droYak2.chr2L 2324486 90 + 22324452 -------------GGCGAAGUGCAGGCAAAGUGGCACU-CGCACUCGG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU--------------- -------------.((((.((((..((...)).)))))-)))((((((-((...((((((.....))))))....(((((.......)))))....)))))))).--------------- ( -38.80, z-score = -4.36, R) >droSec1.super_5 515554 90 + 5866729 -------------GGCGAAGUGCAGGCAAAGUGGCACU-CGCACUCGG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU--------------- -------------.((((.((((..((...)).)))))-)))((((((-((...((((((.....))))))....(((((.......)))))....)))))))).--------------- ( -38.80, z-score = -4.36, R) >droSim1.chr2L 2301404 90 + 22036055 -------------GGCGAAGUGCAGGCAAAGUGGCACU-CGCACUCGG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUAAAGUGCUACCUCGCCGAGUU--------------- -------------.((((.((((..((...)).)))))-)))((((((-(((..((((((.....))))))....(((((.......)))))...))))))))).--------------- ( -40.20, z-score = -5.05, R) >droWil1.scaffold_180708 10241372 104 - 12563649 ---------------GAUAGUGUAGUCAAAGUGGCAUUUCGCUCUCAA-CGAAAGUCACUCGUAAAGUGGCAUUAAGCAUAUUCCAACUGCUCCUGCUGCUGCUUCUAACUGGCUGACUU ---------------...(((.((((((.((((((.(((((.......-))))))))))).....((..(((...((((.........))))..)))..)).........))))))))). ( -27.30, z-score = -1.15, R) >dp4.chr4_group3 7634227 104 - 11692001 UGGGGGGUUCCACGGUGAAGUGCAGGCAAAGUGGCACU-CGCACUCGGGCGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU--------------- .(((((((..(((..(((((((...((..((((.((((-........(((....)))........)))).))))..)).))))))).)))..))))).)).....--------------- ( -41.59, z-score = -2.70, R) >droPer1.super_1 4752435 103 - 10282868 -GGGGGGUUCCACGGUGAAGUGCAGGCAAAGUGGCACU-CGCACUCGGGCGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU--------------- -(((((((..(((..(((((((...((..((((.((((-........(((....)))........)))).))))..)).))))))).)))..))))).)).....--------------- ( -41.59, z-score = -2.94, R) >droVir3.scaffold_12963 13803683 89 + 20206255 -------------CGGGAGCUGCACGCAAAGUGGCACU-CGCACUCAG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGGCAGC-UGCUGAGUU--------------- -------------(((.((((((.(((..((((((..(-(((.....)-)))..))))))....(((((((.....)).)))))...))))))))-).)))....--------------- ( -35.60, z-score = -3.05, R) >droMoj3.scaffold_6500 26495776 89 - 32352404 -------------CGAGAACUGCACGCAAAGUGGCACU-CGCACUCAG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGGCAGC-CGCCGAGUU--------------- -------------((((...(((.(((...))))))))-)).((((.(-((...((((((.(..(((((((.....)).)))))).))))))...-))).)))).--------------- ( -26.60, z-score = -0.91, R) >droGri2.scaffold_15126 2314661 89 - 8399593 -------------CGAGAGCUGCAGCCAAAGUGGUACU-CGCACUCUG-CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGUCAGC-CGCCGAGUU--------------- -------------((((.((..(.......)..)).))-)).((((.(-((...((((((.....)))))).....((((.......))))....-))).)))).--------------- ( -26.70, z-score = -1.14, R) >consensus _____________GGCGAAGUGCAGGCAAAGUGGCACU_CGCACUCGG_CGAAAGUCACUCGUAAAGUGGCAUUAAGCACAUUUCAAGUGCUACCCCGCCGAGUU_______________ ..............................(((......)))((((((.((...((((((.....)))))).....((((.......)))).....))))))))................ (-18.94 = -19.53 + 0.59)
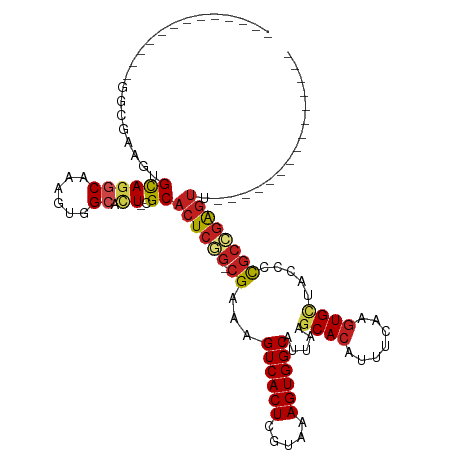
| Location | 2,344,133 – 2,344,223 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.74 |
| Shannon entropy | 0.30511 |
| G+C content | 0.53066 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.64 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2344133 90 - 23011544 ---------------AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CCGAGUGCG-AGUGCCACUUUGCCUGCACUUCGCC------------- ---------------.((((((((..(...(((((((((((.((.....)))).))).))))))..)..))-))))))(((-(((((..........)))).)))).------------- ( -37.80, z-score = -4.00, R) >droAna3.scaffold_12916 10064900 90 + 16180835 ---------------AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CCGAGUGCG-AGUGCCACUUUGCCUGCACUUCUUC------------- ---------------.((((((((..(...(((((((((((.((.....)))).))).))))))..)..))-))))))..(-(((((..........))))))....------------- ( -33.80, z-score = -3.20, R) >droEre2.scaffold_4929 2383216 90 - 26641161 ---------------AACUCGGCGGGGCAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CCGAGUGCG-AGUGCCACUUUGCCUGCACUUCGUC------------- ---------------.((((((((..(...(((((((((((.((.....)))).))).))))))..)..))-))))))(((-(((((..........)))).)))).------------- ( -35.90, z-score = -3.15, R) >droYak2.chr2L 2324486 90 - 22324452 ---------------AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CCGAGUGCG-AGUGCCACUUUGCCUGCACUUCGCC------------- ---------------.((((((((..(...(((((((((((.((.....)))).))).))))))..)..))-))))))(((-(((((..........)))).)))).------------- ( -37.80, z-score = -4.00, R) >droSec1.super_5 515554 90 - 5866729 ---------------AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CCGAGUGCG-AGUGCCACUUUGCCUGCACUUCGCC------------- ---------------.((((((((..(...(((((((((((.((.....)))).))).))))))..)..))-))))))(((-(((((..........)))).)))).------------- ( -37.80, z-score = -4.00, R) >droSim1.chr2L 2301404 90 - 22036055 ---------------AACUCGGCGAGGUAGCACUUUAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CCGAGUGCG-AGUGCCACUUUGCCUGCACUUCGCC------------- ---------------.(((((((((((((((((.......)))))......(((((...))))))).))))-))))))(((-(((((..........)))).)))).------------- ( -36.10, z-score = -4.30, R) >droWil1.scaffold_180708 10241372 104 + 12563649 AAGUCAGCCAGUUAGAAGCAGCAGCAGGAGCAGUUGGAAUAUGCUUAAUGCCACUUUACGAGUGACUUUCG-UUGAGAGCGAAAUGCCACUUUGACUACACUAUC--------------- .(((((........((((..(((....(((((.........)))))..)))..))))..(((((.((((((-(.....)))))).).))))))))))........--------------- ( -25.50, z-score = -0.27, R) >dp4.chr4_group3 7634227 104 + 11692001 ---------------AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCGCCCGAGUGCG-AGUGCCACUUUGCCUGCACUUCACCGUGGAACCCCCCA ---------------.....((.((((((((((.......))))).....((((....((.(((.....))).))(((((.-((.((......))))))))).....)))).))))))). ( -36.90, z-score = -2.59, R) >droPer1.super_1 4752435 103 + 10282868 ---------------AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCGCCCGAGUGCG-AGUGCCACUUUGCCUGCACUUCACCGUGGAACCCCCC- ---------------.....((.((((((((((.......))))).....((((....((.(((.....))).))(((((.-((.((......))))))))).....)))).)))))))- ( -35.20, z-score = -2.24, R) >droVir3.scaffold_12963 13803683 89 - 20206255 ---------------AACUCAGCA-GCUGCCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CUGAGUGCG-AGUGCCACUUUGCGUGCAGCUCCCG------------- ---------------.(((((((.-...(.(((((((((((.((.....)))).))).)))))).)....)-))))))(.(-(((..(((.....)))..)))).).------------- ( -27.50, z-score = -1.37, R) >droMoj3.scaffold_6500 26495776 89 + 32352404 ---------------AACUCGGCG-GCUGCCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CUGAGUGCG-AGUGCCACUUUGCGUGCAGUUCUCG------------- ---------------.((((((((-(..(.(((((((((((.((.....)))).))).)))))).)..)))-)))))).((-((.(((((.....)))..)).))))------------- ( -30.70, z-score = -1.86, R) >droGri2.scaffold_15126 2314661 89 + 8399593 ---------------AACUCGGCG-GCUGACACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG-CAGAGUGCG-AGUACCACUUUGGCUGCAGCUCUCG------------- ---------------......(((-((..((((((((((((.((.....)))).))).))))))...((((-(.....)))-)).......)..)))))........------------- ( -26.20, z-score = -0.86, R) >consensus _______________AACUCGGCGGGGUAGCACUUGAAAUGUGCUUAAUGCCACUUUACGAGUGACUUUCG_CCGAGUGCG_AGUGCCACUUUGCCUGCACUUCGCC_____________ ................((((((((......((((((.((.((((.....).))).)).)))))).....)).))))))(((.((.....)).)))......................... (-17.71 = -18.64 + 0.93)

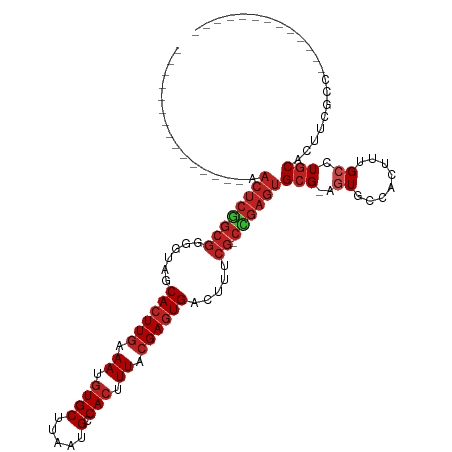
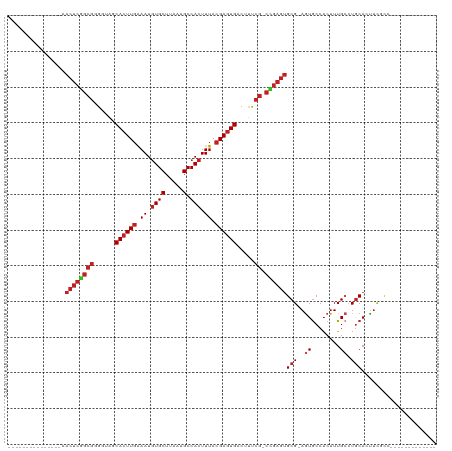
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:59 2011