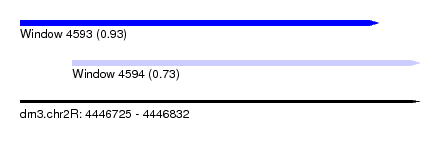
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,446,725 – 4,446,832 |
| Length | 107 |
| Max. P | 0.928789 |

| Location | 4,446,725 – 4,446,821 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.26 |
| Shannon entropy | 0.51702 |
| G+C content | 0.51216 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -11.02 |
| Energy contribution | -10.46 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
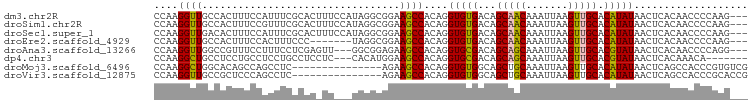
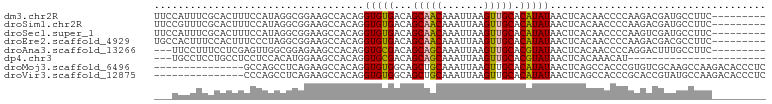
>dm3.chr2R 4446725 96 + 21146708 CCAAGGUUGCCACUUUCCAUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAG--- ....(((((............((((((((......(((....)))..))))))))..(((((.......)))))............))))).....--- ( -26.70, z-score = -3.70, R) >droSim1.chr2R 3087032 96 + 19596830 CCAAGGUUGCCACUUUCCGUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAG--- ....(((((............((((((((......(((....)))..))))))))..(((((.......)))))............))))).....--- ( -26.70, z-score = -3.26, R) >droSec1.super_1 2088580 96 + 14215200 CCAAGGUUGACACUUUCCAUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAG--- ....(((((............((((((((......(((....)))..))))))))..(((((.......)))))............))))).....--- ( -27.40, z-score = -4.03, R) >droEre2.scaffold_4929 8447936 89 - 26641161 CCAAGGUUGCCACUUUCCACUUUCCC-------UAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAG--- ....(((((........(((...((.-------..(((....)))...)).)))...(((((.......)))))............))))).....--- ( -23.90, z-score = -2.78, R) >droAna3.scaffold_13266 18325948 93 - 19884421 CCAAGGUUGGCCGUUUCCUUUCCUCGAGUU---GGCGGAGAAGCCACAGGUGCGACAGCAGCAAAUUAAGUUGCACGUAUAACUCACAACCCCAGG--- ((..((((..(((((..(((.....)))..---)))))...))))....(((((...(((((.......))))).)))))..............))--- ( -24.10, z-score = -0.14, R) >dp4.chr3 2415007 89 + 19779522 CCAAGGCUGCCUCCUGCCUCCUGCCUCCUC---CACAUGGAAGCCACAGGUGCGACAGCAGCAAAUUAAGUUGCACGUAUAACUCACAAACA------- .....(((((.((.(((((...((.(((..---.....))).))...))))).))..)))))......(((((......)))))........------- ( -20.80, z-score = -0.20, R) >droMoj3.scaffold_6496 9367824 84 - 26866924 CCAAGGCUGGCACAGCCAGCCUC---------------AGAAGCCACAGGUGUGGCAGCUGCAAAUUAAGUUGCACAUAUAACUCAGCCACCCGUGUCG ...((((((((...)))))))).---------------....(.(((.((((((((((((........)))))).))...........)))).))).). ( -29.50, z-score = -1.94, R) >droVir3.scaffold_12875 9598162 84 + 20611582 CCAAGGUUGCCGCUCCCAGCCUC---------------AGAAGCCACAGGUGUGGCAGCUGCAAAUUAAGUUGCACAUAUAACUCAGCCACCCGCACCG ...((((((.......)))))).---------------..........(((((((..((((.......(((((......)))))))))...))))))). ( -23.11, z-score = -0.87, R) >consensus CCAAGGUUGCCACUUUCCACUUCGCAC_UU___UAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAG___ ....((((.................................))))....(((((...(((((.......))))).)))))................... (-11.02 = -10.46 + -0.56)
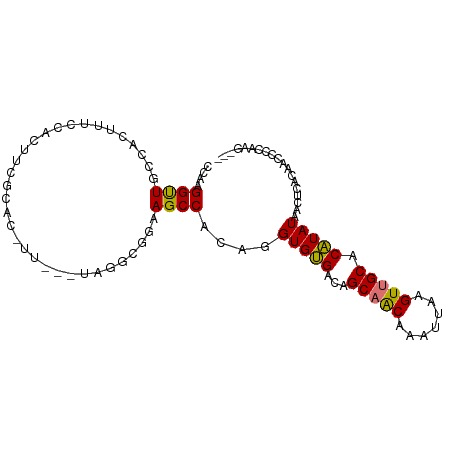
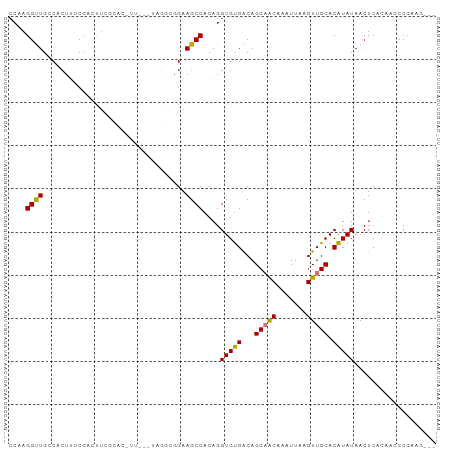
| Location | 4,446,739 – 4,446,832 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.65866 |
| G+C content | 0.50088 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -8.32 |
| Energy contribution | -7.95 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4446739 93 + 21146708 UUCCAUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGAUGCCUUC--------- ...((((((((((((......(((....)))..))))))))..(((((.......)))))........................)))).....--------- ( -21.40, z-score = -2.26, R) >droSim1.chr2R 3087046 93 + 19596830 UUCCGUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGAUGCCUUC--------- ...((((((((((((......(((....)))..))))))))..(((((.......))))).....................))))........--------- ( -23.50, z-score = -2.60, R) >droSec1.super_1 2088594 93 + 14215200 UUCCAUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGUCGAUGCCUUC--------- .......(((.((((......(((....)))...(((((((..(((((.......))))).........)))).)))..))))))).......--------- ( -21.80, z-score = -2.31, R) >droEre2.scaffold_4929 8447943 93 - 26641161 UGCCACUUUCCACUUUCCCUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGACGCCUUC--------- .....................(((....)))..((((((....(((((.......)))))............(........)...))))))..--------- ( -19.20, z-score = -1.24, R) >droAna3.scaffold_13266 18325962 90 - 19884421 ---UUCCUUUCCUCGAGUUGGCGGAGAAGCCACAGGUGCGACAGCAGCAAAUUAAGUUGCACGUAUAACUCACAACCCCAGGACUUUGCCUUC--------- ---.((((......((((((((......)))....(((((...(((((.......))))).))))))))))........))))..........--------- ( -21.44, z-score = -0.24, R) >dp4.chr3 2415021 76 + 19779522 ---UGCCUCCUGCCUCCUCCACAUGGAAGCCACAGGUGCGACAGCAGCAAAUUAAGUUGCACGUAUAACUCACAAACAU----------------------- ---.((((...((.(((.......))).))...))))(((...(((((.......))))).)))...............----------------------- ( -16.50, z-score = -0.71, R) >droMoj3.scaffold_6496 9367838 87 - 26866924 ---------------GCCAGCCUCAGAAGCCACAGGUGUGGCAGCUGCAAAUUAAGUUGCACAUAUAACUCAGCCACCCGUGUCGCAAGCCAAGACACCCUC ---------------....(.((.((..(((((....))))).(.(((((......))))))......)).)).)....(((((.........))))).... ( -19.60, z-score = 0.04, R) >droVir3.scaffold_12875 9598176 87 + 20611582 ---------------CCCAGCCUCAGAAGCCACAGGUGUGGCAGCUGCAAAUUAAGUUGCACAUAUAACUCAGCCACCCGCACCGUAUGCCAAGACACCCUC ---------------.............((....(((((((..((((.......(((((......)))))))))...)))))))....))............ ( -19.11, z-score = -0.43, R) >consensus ___CACUUCGCACUUUCCAUACGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGAUGCCUUC_________ ...................................(((((...(((((.......))))).))))).................................... ( -8.32 = -7.95 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:20 2011