| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,436,574 – 4,436,667 |
| Length | 93 |
| Max. P | 0.879276 |

| Location | 4,436,574 – 4,436,667 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 68.30 |
| Shannon entropy | 0.59972 |
| G+C content | 0.40432 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -8.67 |
| Energy contribution | -8.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879276 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 4436574 93 + 21146708 AUCUACGUAAUUUACGCACAAGCAUUU------CGUUCCC---GACUAUGUGGGCGAUUUUCC-UUUGCUUCGCCCAUAAGUUUAUUUAUGUUUAU-UUUGUGG .....((((...))))((((((.....------(((....---((((.(((((((((......-......))))))))))))).....))).....-)))))). ( -23.10, z-score = -3.12, R) >droSim1.chr2R 3075378 89 + 19596830 AUCUACGUAAUUUACGCACAAGCAUUU------CGUUCCCUCUGACUAUGUGGGCGAUUUUCC-UCUGCUUCGCCCAUAAGUU-------GUUUAU-UUUGUGG ..(((((.(((..((((....))....------..........((((.(((((((((......-......)))))))))))))-------))..))-).))))) ( -23.40, z-score = -3.43, R) >droSec1.super_1 2078471 89 + 14215200 AUCUACGUAAUUUACGCACAAGCAUUU------CGUUCCCUAUGACUAUGUGGGCGAUUUUCC-UCUGCUUCGCCCAUAAGUU-------GUUUAU-UUUGUGG ..(((((.(((..((((....))....------..........((((.(((((((((......-......)))))))))))))-------))..))-).))))) ( -23.40, z-score = -3.27, R) >droEre2.scaffold_4929 8437643 86 - 26641161 AUCUACGUAAUUUACGCACAAGCAUUU------CAUUCCG---GACUAUGUGGGCAAUUUUCC-UCAGCUUUAUCUAUAAGUU-------GUUUAU-UUUGUGG ..(((((((......((....))..((------(.....)---)).)))))))((((......-.((((((.......)))))-------).....-.)))).. ( -13.42, z-score = -0.43, R) >droYak2.chr2L 17092411 86 + 22324452 AUCUACGUAAUUUACGCACAAGGAUUU------CAUUUCG---GACUAUGUGGGCCAUUUUCC-UCGGCUUUAUCCAUAAGUU-------GUUUAU-UUUGUGG .....((((...))))((((((((...------...))).---((((.((((((((.......-..))).....)))))))))-------......-.))))). ( -14.60, z-score = -0.12, R) >droAna3.scaffold_13266 18314765 78 - 19884421 AUCUACGUAAUUUACGCAUACAUAUUU------GAGCUCA---GAGGAUGUGGGUG---------UCCUUGCUCCCGACAGUU-------GUUUAU-UUUUCGG ......((((...((((.(((((.(((------(....))---))..))))).)))---------)..))))..((((.((..-------......-)).)))) ( -15.00, z-score = 0.09, R) >dp4.chr3 2405899 83 + 19779522 AUCUACGUAAUUUAUGCACACAC---U------GAGA-CA---GACGAUGUGGGCGAACGGCUGCUCUCCUCGCCCAUAAAUU-------GUUUAU-UUUGCGG .....(((((..((.(((....(---(------(...-))---)....(((((((((..((.......)))))))))))...)-------)).)).-.))))). ( -22.40, z-score = -1.89, R) >droPer1.super_2 2595455 83 + 9036312 AUCUACGUAAUUUAUGCACACAC---U------GAGA-CA---GACGAUGUGGGCGAACGGCUGCUCUCCUCGCCCAUAAAUU-------GUUUAU-UUUGCGG .....(((((..((.(((....(---(------(...-))---)....(((((((((..((.......)))))))))))...)-------)).)).-.))))). ( -22.40, z-score = -1.89, R) >droWil1.scaffold_180700 3595535 96 - 6630534 AUUUAUGUAAUUUAUGCAAAAGUCCCUUUACAACAAUGUGUUUUACCAUGUGUGUGUGUG-----UGUGUAUAACCAUAAAUUGUUU---AUAUGUGUUUGCAG ..............((((((.........((((..(((.(((.(((((..(......)..-----)).))).))))))...))))..---.......)))))). ( -15.57, z-score = 0.37, R) >consensus AUCUACGUAAUUUACGCACAAGCAUUU______CAUUCCA___GACUAUGUGGGCGAUUUUCC_UUCGCUUCGCCCAUAAGUU_______GUUUAU_UUUGUGG .....((((...))))................................(((((((((.............)))))))))......................... ( -8.67 = -8.48 + -0.19)

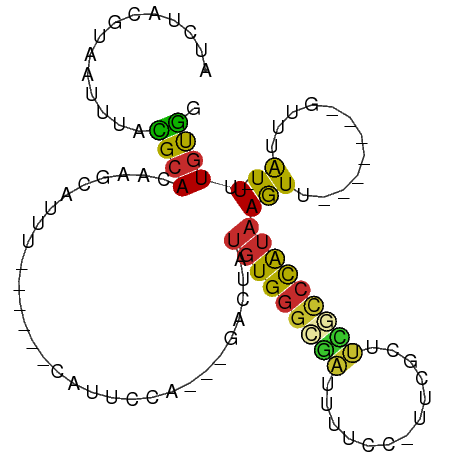

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:18 2011