
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,383,023 – 4,383,134 |
| Length | 111 |
| Max. P | 0.603188 |

| Location | 4,383,023 – 4,383,134 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.51688 |
| G+C content | 0.37711 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -9.12 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
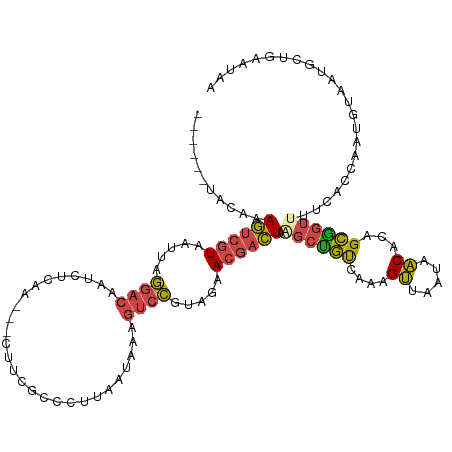
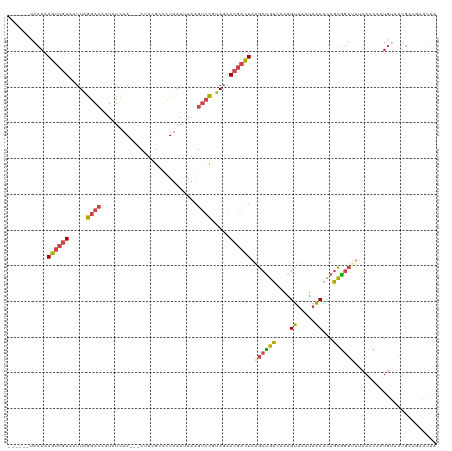
>dm3.chr2R 4383023 111 + 21146708 ------UACAAAGUCGUAAUUAGGACAAUCUCAA---CUUCGCCCUUAAUAAAGUCCGUAGAACGACUAGGCUGUCAAAGUUAAUAACACAGCGGUUGUCACCAAUGUAAUGCUGAAUAA ------((((.((((((.....((((........---................)))).....))))))..(((((.............)))))(((....)))..))))........... ( -19.98, z-score = -0.37, R) >droSim1.chr2R 2996015 111 + 19596830 ------UACAAAGUCGUAAUUAGGACAAUCUCAA---CUUCGCCCUUAAUAAAGUCCGUAGAACGACUAAGCUGUCAAAGUUAAUAACACAGCGGUUUUCAUCAGUGUAAUGCUGAAUAA ------.....((((((.....((((........---................)))).....))))))..(((((.............)))))........(((((.....))))).... ( -21.48, z-score = -1.39, R) >droSec1.super_1 2024818 111 + 14215200 ------UACAAAGUCGUAAUUAGGACAAUCUCAA---CUUCGCCCUUAAUAAAGUCCGUAGAACGACUAAGCUGUCAAAGUUAAUAACACAGCGGUUUUCAUCAGUGUAAUGCUGAAUAA ------.....((((((.....((((........---................)))).....))))))..(((((.............)))))........(((((.....))))).... ( -21.48, z-score = -1.39, R) >droYak2.chr2L 17036839 111 + 22324452 ------UACAAAGUCGUAAUUAGGACAAUCUCAA---CUUCGCCCUUAAUAAAGUCCUUAGAACGACUAAGCUGUCAAAGUUAAUAACACAGCGGUAUUCACCAAUGUUAUGUGAAAUAA ------((((.((((((....(((((........---................)))))....))))))..(((((.............)))))(((....))).......))))...... ( -21.28, z-score = -1.90, R) >droEre2.scaffold_4929 8383768 102 - 26641161 ------CACAAAGUCGUAAUUAGGAGCAUCUCAA---CUUCGCCUUUAAUAA-GUCCUUAGAAGGACUAAGCCGCUAAGGUUAAUAACACAGCGGUUUUCAGCAAUGGCAUG-------- ------......(((((.....((((........---))))((........(-(((((....))))))((((((((...((.....))..))))))))...)).)))))...-------- ( -26.00, z-score = -2.38, R) >droPer1.super_2 2528652 99 + 9036312 GUUUACUACAAAA--GUAGAUCAGACACUUUUGAGAGCUAAGCUCUAAAUAGAGCAUGGAAAACCUUUGAUAUUCCUGGGGAAUCAUCAAAGAAA-----AGCGAU-------------- .....((.(((((--((.(......)))))))))).(((..(((((....))))).........((((((((((((...))))).)))))))...-----)))...-------------- ( -25.90, z-score = -2.97, R) >consensus ______UACAAAGUCGUAAUUAGGACAAUCUCAA___CUUCGCCCUUAAUAAAGUCCGUAGAACGACUAAGCUGUCAAAGUUAAUAACACAGCGGUUUUCACCAAUGUAAUGCUGAAUAA ...........((((((.....((((...........................)))).....)))))).((((((....((.....))...))))))....................... ( -9.12 = -9.10 + -0.02)

| Location | 4,383,023 – 4,383,134 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.51688 |
| G+C content | 0.37711 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -11.91 |
| Energy contribution | -11.34 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
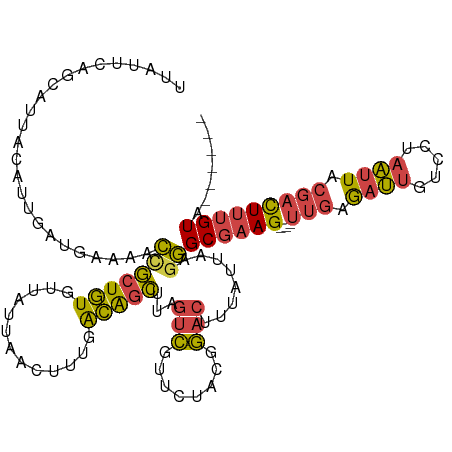
>dm3.chr2R 4383023 111 - 21146708 UUAUUCAGCAUUACAUUGGUGACAACCGCUGUGUUAUUAACUUUGACAGCCUAGUCGUUCUACGGACUUUAUUAAGGGCGAAG---UUGAGAUUGUCCUAAUUACGACUUUGUA------ .....((((......(((....)))..))))(((((.......)))))((..((((((((....))........(((((((..---......)))))))....))))))..)).------ ( -25.30, z-score = -0.85, R) >droSim1.chr2R 2996015 111 - 19596830 UUAUUCAGCAUUACACUGAUGAAAACCGCUGUGUUAUUAACUUUGACAGCUUAGUCGUUCUACGGACUUUAUUAAGGGCGAAG---UUGAGAUUGUCCUAAUUACGACUUUGUA------ ....((((.......))))......(((((((.............)))))..((((........)))).......))((((((---(((.(((((...))))).))))))))).------ ( -22.92, z-score = -0.51, R) >droSec1.super_1 2024818 111 - 14215200 UUAUUCAGCAUUACACUGAUGAAAACCGCUGUGUUAUUAACUUUGACAGCUUAGUCGUUCUACGGACUUUAUUAAGGGCGAAG---UUGAGAUUGUCCUAAUUACGACUUUGUA------ ....((((.......))))......(((((((.............)))))..((((........)))).......))((((((---(((.(((((...))))).))))))))).------ ( -22.92, z-score = -0.51, R) >droYak2.chr2L 17036839 111 - 22324452 UUAUUUCACAUAACAUUGGUGAAUACCGCUGUGUUAUUAACUUUGACAGCUUAGUCGUUCUAAGGACUUUAUUAAGGGCGAAG---UUGAGAUUGUCCUAAUUACGACUUUGUA------ .........(((((((.((((.....)))))))))))...........((..((((((....(((((....((((........---))))....)))))....))))))..)).------ ( -24.30, z-score = -1.01, R) >droEre2.scaffold_4929 8383768 102 + 26641161 --------CAUGCCAUUGCUGAAAACCGCUGUGUUAUUAACCUUAGCGGCUUAGUCCUUCUAAGGAC-UUAUUAAAGGCGAAG---UUGAGAUGCUCCUAAUUACGACUUUGUG------ --------...(((...........((((((.((.....))..))))))...((((((....)))))-).......)))((((---(((.(((.......))).)))))))...------ ( -26.70, z-score = -2.16, R) >droPer1.super_2 2528652 99 - 9036312 --------------AUCGCU-----UUUCUUUGAUGAUUCCCCAGGAAUAUCAAAGGUUUUCCAUGCUCUAUUUAGAGCUUAGCUCUCAAAAGUGUCUGAUCUAC--UUUUGUAGUAAAC --------------...(((-----..(((((((((.((((...)))))))))))))........(((((....)))))..))).((((((((((.......)))--))))).))..... ( -25.00, z-score = -2.93, R) >consensus UUAUUCAGCAUUACAUUGAUGAAAACCGCUGUGUUAUUAACUUUGACAGCUUAGUCGUUCUACGGACUUUAUUAAGGGCGAAG___UUGAGAUUGUCCUAAUUACGACUUUGUA______ ...........................(((((.............)))))..((((((.....((((....((((...........))))....)))).....))))))........... (-11.91 = -11.34 + -0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:08 2011