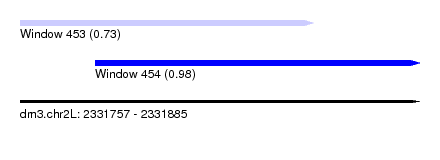
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,331,757 – 2,331,885 |
| Length | 128 |
| Max. P | 0.976914 |

| Location | 2,331,757 – 2,331,851 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 62.29 |
| Shannon entropy | 0.71954 |
| G+C content | 0.49115 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.28 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2331757 94 + 23011544 ---ACUGCUUCAUCAGCGGGUUUU---GAGCGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------CUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG ---.......((((.(((((....---((((((((((.((.(((.(.(((((......))))))))--------).)).)))))---)))))......)))))....)))) ( -33.70, z-score = -2.66, R) >droSim1.chr2L 2289205 94 + 22036055 ---ACUGCUUCAUCAGCGGGUUUU---GAGCAAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------CUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG ---.......((((.(((((....---((((.(((((.((.(((.(.(((((......))))))))--------).)).)))))---.))))......)))))....)))) ( -27.80, z-score = -0.99, R) >droSec1.super_5 503378 94 + 5866729 ---ACUGCUUCAUCAGCGGGUUUU---GAGCGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------CUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG ---.......((((.(((((....---((((((((((.((.(((.(.(((((......))))))))--------).)).)))))---)))))......)))))....)))) ( -33.70, z-score = -2.66, R) >droYak2.chr2L 2311239 102 + 22324452 ---ACUGCUUCAUCAGCGGGUUUU---GAGCGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUGUUCACCUGCUCACCUGUU---CGCUCUACUUUUUCGCGCUCGGUG ---.......((((((((.(....---((((((((((..(((.(((.(((((......))))).....))).)))....)))))---)))))........).)))).)))) ( -36.90, z-score = -3.05, R) >droEre2.scaffold_4929 2370751 94 + 26641161 ---ACUGCUUCAUCAGCGGGUUUU---GAGCGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------UUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG ---.......((((.(((((....---((((((((((.((.....(.(((((......))))))..--------..)).)))))---)))))......)))))....)))) ( -31.00, z-score = -1.99, R) >droAna3.scaffold_12916 10052100 100 - 16180835 ---ACUGCUUCAUCAGCGGUUUUUUUAGAGCGAGCAGGUGCGUGUGUGUGCAUAAAUUUGCACCUG--------UUCGCCGGUGUGGCGUUCCAAUUUCCACCGCUCGCUG ---..........((((((..........(((((((((((((((((....))))....))))))))--------)))))((((((((....))).....))))).)))))) ( -39.10, z-score = -3.08, R) >dp4.chr4_group3 7613726 89 - 11692001 ---UCUGCUUCAUCAGCGGCUUUU---GAGCG-GUGUGUGCGUGUGUGUGCAUAAAUUUGCACCUC--------UUCAAAAGGAGUUCCUCCGCCUCCUCCACA------- ---...(((.....)))(((((((---(((.(-(((((....((((....))))....)))))).)--------.))))))((((...))))))).........------- ( -25.70, z-score = -1.39, R) >droGri2.scaffold_15126 2300039 98 - 8399593 UUUGCACUCUUAUCAUCAGGUGAAUAUGAAUAUGAAAAUAUGAUUGGAAAUUCAAGUGAAUGUAGG--------CCCCACUCCA---CCCUAUAUUUCUCCAUAUUUCU-- .........(((((....)))))....(((((((......(((.(....).)))...(((((((((--------..........---.)))))))))...)))))))..-- ( -13.40, z-score = 1.02, R) >droVir3.scaffold_12963 13786504 90 + 20206255 UUUGCACCUUUAUCAUCAGGUAAAUAUGAAUAUGCAAAUAUGAUUGGGAAU----GUGAGCGUACU--------CACUCCGCCA---CCCGCUGCCUUCACUUCU------ .....((((........)))).....((((...(((..........(((..----(((((....))--------))))))((..---...))))).)))).....------ ( -14.80, z-score = 0.61, R) >consensus ___ACUGCUUCAUCAGCGGGUUUU___GAGCGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG________CUCACCUGUU___CGCUCUACUUUUUCGCGUUCGGUG ....(((((.....)))))........(((((.(((....)))....(((((......))))).......................................))))).... ( -8.93 = -9.28 + 0.35)

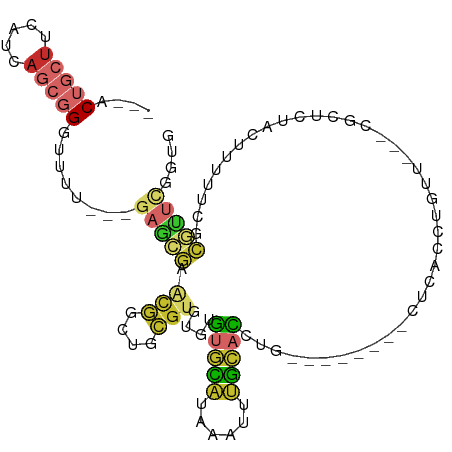
| Location | 2,331,781 – 2,331,885 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 61.77 |
| Shannon entropy | 0.74228 |
| G+C content | 0.49757 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.04 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2331781 104 + 23011544 CGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------CUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG-----GCAAUUUGCACACAUUUUCACGUAGCCAGGGAGA- .....(((((((((((((((((............((--------(.((((.(..---(((...........)))..)))))-----)))...))))))))....))))))))).......- ( -38.76, z-score = -2.89, R) >droSim1.chr2L 2289229 104 + 22036055 CAAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------CUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG-----GCAAUUUGCACACAUUUUCACGUAGCCAGGGAGA- .....(((((((((((((((((............((--------(.((((.(..---(((...........)))..)))))-----)))...))))))))....))))))))).......- ( -38.76, z-score = -3.19, R) >droSec1.super_5 503402 104 + 5866729 CGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------CUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG-----GCAAUUUGCACACAUUUUCACGUAGCCAGGGAGA- .....(((((((((((((((((............((--------(.((((.(..---(((...........)))..)))))-----)))...))))))))....))))))))).......- ( -38.76, z-score = -2.89, R) >droYak2.chr2L 2311263 112 + 22324452 CGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUGUUCACCUGCUCACCUGUU---CGCUCUACUUUUUCGCGCUCGGUG-----GCAAUUUGCACACAUUUUCACGUAGCCAGGGAGA- .....(((((((((((((((((.....((((....((......)).((((.(..---(((...........)))..)))))-----))))..))))))))....))))))))).......- ( -39.90, z-score = -2.53, R) >droEre2.scaffold_4929 2370775 104 + 26641161 CGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG--------UUCACCUGUU---CGCUCUACUUUUUCGCGUUCGGUG-----GCAAUUUGCACACAUUUCCACGUAGCCAGGGAGA- .....(((((((((((((((((.....((((.....--------..((((.(..---(((...........)))..)))))-----))))..))))))))....))))))))).......- ( -38.51, z-score = -2.88, R) >droAna3.scaffold_12916 10052127 107 - 16180835 CGAGCAGGUGCGUGUGUGUGCAUAAAUUUGCACCUG--------UUCGCCGGUGUGGCGUUCCAAUUUCCACCGCUCGCUG-----GCAAUUUGCAAGCAUUUCCACGUAGUCGUGGCGA- .(((((((((((((((....))))....))))))))--------)))(((((((.((((.............)))))))))-----))...............(((((....)))))...- ( -42.92, z-score = -2.31, R) >dp4.chr4_group3 7613750 99 - 11692001 -CGGUGUGUGCGUGUGUGUGCAUAAAUUUGCACCUC--------UUCAAAAGGA----GUUCCUCCGCCUC---CUCCACA-----GCUGAUUGCACGCCUACCCCCGUCCAUCGGGGGG- -.(((((((((.((((.(((((......)))))...--------......((((----(.........)))---)).))))-----)).....)))))))..((((((.....)))))).- ( -39.40, z-score = -3.51, R) >droGri2.scaffold_15126 2300063 109 - 8399593 --------UAUGAAUAUGAAAAUAUGAUUGGAAAUU----CAAGUGAAUGUAGGCCCCACUCCACCCUAUAUUUCUCCAUAUUUCUGUUCCUCGUAUGCAUUUUAACGUAUUCAUUGCACA --------.(((((((((...((((((.(((((((.----.....(((((((((...........)))))))))......)))))))....)))))).........)))))))))...... ( -20.00, z-score = -1.34, R) >droVir3.scaffold_12963 13786528 101 + 20206255 --------UAUGAAUAUGCAAAUAUGAUUGGGAAU--------GUGAGCGUA---CUCACUCCGCCACCCGCUGC-CUUCACUUCUGUUUCUCGUAUGCAUUUUCACGUACUCAAUGCACA --------..((((.(((((..(((((...(((..--------(((((....---))))))))((.....))...-...............)))))))))).))))............... ( -19.80, z-score = -0.20, R) >consensus CGAACGGCUGCGUGUGUGUGCAUAAAUUUGCACCUG________UUCACCUGUU___CGCUCUACUUUUUCGCGCUCGGUG_____GCAAUUUGCACACAUUUUCACGUAGCCAGGGAGA_ ......((((((.(((((((((......................................................................))))))))).....))))))......... ( -9.86 = -10.04 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:57 2011