
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,354,839 – 4,354,941 |
| Length | 102 |
| Max. P | 0.566373 |

| Location | 4,354,839 – 4,354,941 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.96 |
| Shannon entropy | 0.62925 |
| G+C content | 0.45100 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
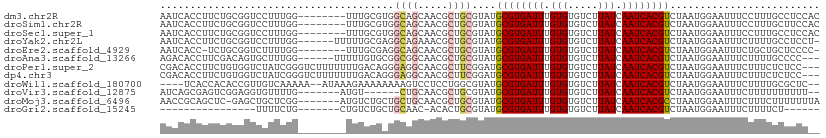
>dm3.chr2R 4354839 102 - 21146708 AAUCACCUUCUGCGGUCCUUUGG--------UUUGCGUGGCAGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCCUUUGCCUCCAC .....((......))(((.((((--------..((((..((......))..)))).((((((((.(((.....))).)))))))))))).)))................. ( -25.30, z-score = -0.80, R) >droSim1.chr2R 2985476 102 - 19596830 AAUCACCUUCUGCGGUCCUUUGG--------UUUGCGUGGCAGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCCUUUGCUUCCAC ...........(((((((.((((--------..((((..((......))..)))).((((((((.(((.....))).)))))))))))).))).......))))...... ( -25.31, z-score = -0.87, R) >droSec1.super_1 1996892 102 - 14215200 AAUCACCUUCUGCGGUCCUUUGG--------UUUGCGUGGCAGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCCUUUGCCUCCAC .....((......))(((.((((--------..((((..((......))..)))).((((((((.(((.....))).)))))))))))).)))................. ( -25.30, z-score = -0.80, R) >droYak2.chr2L 17009875 103 - 22324452 AAUCACCUUCUGCGGUCCUUUGG------UUUUUGCGAGGCAGAAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUUGCCUCCU- ...........((((.((...))------...))))((((((((((..((...((.((((((((.(((.....))).)))))))).))..))..))))...))))))..- ( -25.90, z-score = -1.27, R) >droEre2.scaffold_4929 8357513 100 + 26641161 AAUCACC-UCUGCGGUCUUUUGG--------UUUGCGAGGCAGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUGCUGCUCCCC- ......(-((.((((.(.....)--------.)))))))((((((....(.(((..((((((((.(((.....))).))))))))...))).).....)))))).....- ( -25.00, z-score = -0.27, R) >droAna3.scaffold_13266 18234952 101 + 19884421 AGACACCUUCGACAGUGCUUUGG------UUUUUGUGCGGCGGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUUGCCCC--- ................((...((------.(((..(((((((....)))))).((.((((((((.(((.....))).)))))))).)))..)))...))...))...--- ( -27.20, z-score = -0.67, R) >droPer1.super_2 2504581 107 - 9036312 CGACACCUUCUGUGGUCUAUCGGGUCUUUUUUUGACAGGGAGGCAACGCUUCGGAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUCUCUCC--- (((.(((......)))...))).(((.......))).((((((......((((...((((((((.(((.....))).))))))))....)))).......)))))).--- ( -23.42, z-score = 0.43, R) >dp4.chr3 2332851 107 - 19779522 CGACACCUUCUGUGGUCUAUCGGGUCUUUUUUUGACAGGGAGGCAACGCUUCGGAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUCUCUCC--- (((.(((......)))...))).(((.......))).((((((......((((...((((((((.(((.....))).))))))))....)))).......)))))).--- ( -23.42, z-score = 0.43, R) >droWil1.scaffold_180700 3492855 102 + 6630534 ----UCACCACACCGUUGUCAAAAA--AUAAAGAAAAAAAAGUCCUCCUGGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUUGCGCUC-- ----........((((((.......--..............(.((....)))....((((((((.(((.....))).)))))))).))))))................-- ( -16.00, z-score = -0.05, R) >droVir3.scaffold_12875 9425991 95 - 20611582 AUCAGCGAGUCGGAGGUGUUUUG-------AUGU------CUGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUUUUUUUU-- ..(((((...((((.((......-------)).)------)))...)))))(((..((((((((.(((.....))).))))))))...))).................-- ( -21.90, z-score = -0.52, R) >droMoj3.scaffold_6496 9189181 102 + 26866924 AACCGCAGCUC-GAGCUGCUCGG-------AUGUCUGCUGCUGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGCCUAAUGGAAUUUCUUUCUUUUUUUA ..(((((((((-(((...)))))-------..((.(((....))))))))))....((((((((.(((.....))).)))))))).....)).................. ( -29.40, z-score = -1.89, R) >droGri2.scaffold_15245 2691598 80 - 18325388 ----------------UUUUCUG-------CUGUCUGCUGCAAC-ACACUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUUCU------ ----------------..(((((-------(.((.((......)-).)).)).((.((((((((.(((.....))).)))))))).))..))))..........------ ( -14.40, z-score = -0.64, R) >consensus AAUCACCUUCUGCGGUCCUUUGG________UUUGCGAGGCAGCAACGCUGCGUAUGCGUGAUUUGUGUGUCUUAUCAAUCACGUCUAAUGGAAUUUCUUUUGCUUCC__ .......................................((((.....))))....((((((((.(((.....))).))))))))......................... (-12.52 = -12.73 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:04 2011