| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,332,483 – 4,332,583 |
| Length | 100 |
| Max. P | 0.997847 |

| Location | 4,332,483 – 4,332,583 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.45 |
| Shannon entropy | 0.82321 |
| G+C content | 0.43426 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -3.77 |
| Energy contribution | -3.27 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.54 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.12 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
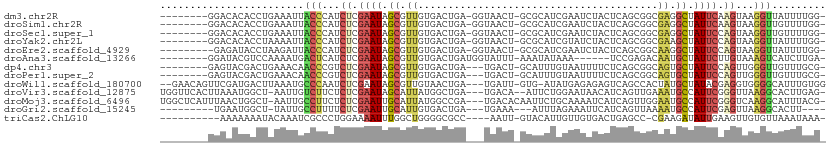
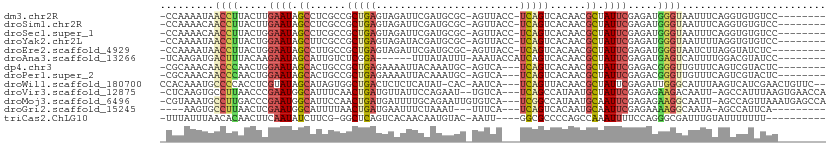
>dm3.chr2R 4332483 100 + 21146708 --------GGACACACCUGAAAUUACCCAUCUCGAAUAGCGUUGUGACUGA-GGUAACU-GCGCAUCGAAUCUACUCAGCGGCGAGGCUAUUCAAGUAAGGUUAUUUUGG- --------........(..((((.(((...((.(((((((.((((..((((-(......-.((...))......)))))..)))).))))))).))...))).))))..)- ( -30.62, z-score = -2.33, R) >droSim1.chr2R 2956859 100 + 19596830 --------GGACACACCUGAAAUUACCCAUCUCGAAUAGCGUUGUGACUGA-GGUAACU-GCGCAUCGAAUCUACUCAGCGGCGAGGCUAUUCAAGUAAGGUUGUUUUGG- --------........(..((((.(((...((.(((((((.((((..((((-(......-.((...))......)))))..)))).))))))).))...))).))))..)- ( -31.02, z-score = -2.04, R) >droSec1.super_1 1974085 100 + 14215200 --------GGACACACCUGAAAUUACCCAUCUCGAAUAGCGUUGUGACUGA-GGUAACU-GCGCAUCGAAUCUACUCAGCGGCGAGGCUAUUCCAGUAAGGUUGUUUUGG- --------........(..((((.(((...((.(((((((.((((..((((-(......-.((...))......)))))..)))).))))))).))...))).))))..)- ( -31.02, z-score = -1.86, R) >droYak2.chr2L 16988559 100 + 22324452 --------GGACACACCUAAAAUUACCCAUCUCGAAUAGCGUUGUGACUGA-GGUAACU-GCGCAUCGUAUCUACUCAGCGGCGAAGCUAUUCCAGUAAGGUUAUUUUGG- --------........(((((((.(((...((.(((((((.((((..((((-(.....(-(((...))))....)))))..)))).))))))).))...))).)))))))- ( -31.10, z-score = -2.87, R) >droEre2.scaffold_4929 8335343 99 - 26641161 ---------GAGAUACCUAAGAUUACCCAUCUCGAAUAGCGUUGUGACUGA-GGUAACU-GCGCAUCGAAUCUACUCAGCGGCAAGGCUAUUCCAGUAAGGUUAUUUUGG- ---------.......(((((((.(((...((.(((((((.((((..((((-(......-.((...))......)))))..)))).))))))).))...))).)))))))- ( -31.52, z-score = -2.78, R) >droAna3.scaffold_13266 18214132 95 - 19884421 --------GGAUACGUCCAAAAUGACUCAUCUCGAAUAGCGUUGUGACUGAUGGUAUUU-AAAUAUAAA------UCCGAGACAAUGCUAUUCUUGUAAAGUCAUCUUGA- --------.........(((.((((((...(..((((((((((((..(.(((..(((..-....))).)------)).)..))))))))))))..)...)))))).))).- ( -26.80, z-score = -3.62, R) >dp4.chr3 2308612 98 + 19779522 --------GAGUACGACUGAAACAACCCGUCUCGAAUAGCGUUGUGACUGA---UGACU-GCAUUUGUAAUUUUCUCAGCGGCAGUGCUAUUCCAGUUGGGUUGUUUGCG- --------...........((((((((((.((.((((((((((((..((((---.((.(-((....))).))...))))..)))))))))))).)).))))))))))...- ( -35.50, z-score = -3.53, R) >droPer1.super_2 2480070 98 + 9036312 --------GAGUACGACUGAAACAACCCGUCUCGAAUAGCGUUGUGACUGA---UGACU-GCAUUUGUAAUUUUCUCAGCGGCAGUGCUAUUCCAGUUGGGUUGUUUGCG- --------...........((((((((((.((.((((((((((((..((((---.((.(-((....))).))...))))..)))))))))))).)).))))))))))...- ( -35.50, z-score = -3.53, R) >droWil1.scaffold_180700 3469396 104 - 6630534 --GAACAGUUCGAUGACUUAAAUGCCCAAUCUCGAAUAGCGUUGUAACUGA---UGAUU-GUG-AUAUGAGAGAGUCAGCCACUAUGCUAUACGAGGUGGGGCAUUUGUGG --..............(.(((((((((.((((((.(((((((.((..((((---(..((-...-......))..)))))..)).))))))).)))))).))))))))).). ( -37.40, z-score = -4.22, R) >droVir3.scaffold_12875 9403394 104 + 20611582 UGGUUCACUUAAAUGGCU-AAUUGUCUUCUCUCGAAUAGCAUUAUGGCUGA---UGACA--AUUCUGGAAUAACAUCAGUUGAAAUGCCAUUCGGGUUAAGGCACUUGAG- ((((.((......)))))-)..((((((..(((((((.(((((.(((((((---((...--(((....)))..))))))))).))))).)))))))..))))))......- ( -31.10, z-score = -3.21, R) >droMoj3.scaffold_6496 9158927 106 - 26866924 UGGCUCAUUUAACUGGCU-AAUUGCCUUCUCUCGAAUUGCAUUAUGGCCGA---UGACACAAUUCUGCAAAAUCAUCAGUUGGAAUGCCAUUCGGGUCAAGGCAUUUACG- ((((.((......)))))-)..((((((..(((((((.(((((.((((.((---(((...............))))).)))).))))).)))))))..))))))......- ( -32.86, z-score = -3.21, R) >droGri2.scaffold_15245 2671305 91 + 18325388 ---------UGAAUGGCU-UAUUGCCUUUUCUCGAAUUGCAUUGUGACUGA---UGAAA---AUUUAGAAAUUCAUCAGUUAAAAUGCCAUUCGAGUUAAGGCACUU---- ---------.........-...((((((..(((((((.(((((.(((((((---((((.---.........))))))))))).))))).)))))))..))))))...---- ( -39.10, z-score = -7.98, R) >triCas2.ChLG10 4636082 94 - 8806720 ----------AAAAAAAUACAAAUCGCCCUGGAAAAUUUGGCUGGGGCGCC----AAUU-GUACAUUGUUGUGACUGAGCC-CGAAGAUAUUGAAGUUGUGUUAAAUAAA- ----------.....(((((((.((((((..(.........)..))))(((----(...-.((((....))))..)).)).-..........))..))))))).......- ( -20.10, z-score = -0.76, R) >consensus ________GGAAACGCCUGAAAUUACCCAUCUCGAAUAGCGUUGUGACUGA___UAACU_GCGCAUCGAAUCUACUCAGCGGCAAUGCUAUUCCAGUAAGGUUAUUUUGG_ ........................(((...((.((((.((.((........................................)).)).)))).))...)))......... ( -3.77 = -3.27 + -0.50)
| Location | 4,332,483 – 4,332,583 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 61.45 |
| Shannon entropy | 0.82321 |
| G+C content | 0.43426 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -7.34 |
| Energy contribution | -6.98 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.997847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
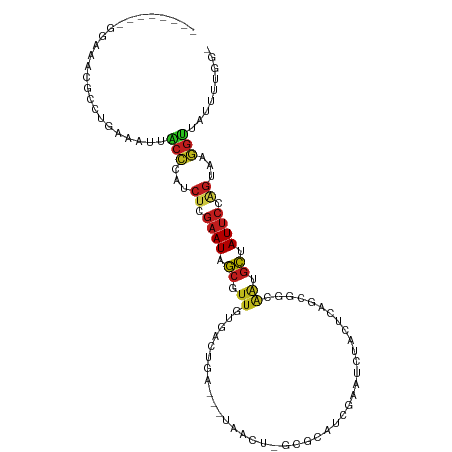
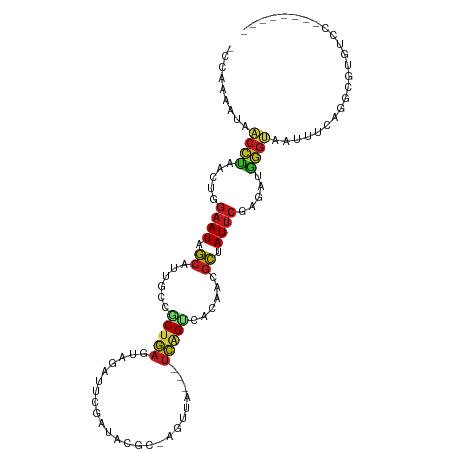
>dm3.chr2R 4332483 100 - 21146708 -CCAAAAUAACCUUACUUGAAUAGCCUCGCCGCUGAGUAGAUUCGAUGCGC-AGUUACC-UCAGUCACAACGCUAUUCGAGAUGGGUAAUUUCAGGUGUGUCC-------- -((.((((.((((..((((((((((......((((((..((((((...)).-))))..)-)))))......))))))))))..)))).))))..)).......-------- ( -28.40, z-score = -2.18, R) >droSim1.chr2R 2956859 100 - 19596830 -CCAAAACAACCUUACUUGAAUAGCCUCGCCGCUGAGUAGAUUCGAUGCGC-AGUUACC-UCAGUCACAACGCUAUUCGAGAUGGGUAAUUUCAGGUGUGUCC-------- -........((((..((((((((((......((((((..((((((...)).-))))..)-)))))......))))))))))..))))................-------- ( -26.70, z-score = -1.50, R) >droSec1.super_1 1974085 100 - 14215200 -CCAAAACAACCUUACUGGAAUAGCCUCGCCGCUGAGUAGAUUCGAUGCGC-AGUUACC-UCAGUCACAACGCUAUUCGAGAUGGGUAAUUUCAGGUGUGUCC-------- -.....(((((((.....(((((((......((((((..((((((...)).-))))..)-)))))......)))))))(((((.....))))))))).)))..-------- ( -25.20, z-score = -0.60, R) >droYak2.chr2L 16988559 100 - 22324452 -CCAAAAUAACCUUACUGGAAUAGCUUCGCCGCUGAGUAGAUACGAUGCGC-AGUUACC-UCAGUCACAACGCUAUUCGAGAUGGGUAAUUUUAGGUGUGUCC-------- -(((((((.((((..((.(((((((......((((((.....((.......-.))...)-)))))......))))))).))..)))).))))).)).......-------- ( -27.20, z-score = -1.49, R) >droEre2.scaffold_4929 8335343 99 + 26641161 -CCAAAAUAACCUUACUGGAAUAGCCUUGCCGCUGAGUAGAUUCGAUGCGC-AGUUACC-UCAGUCACAACGCUAUUCGAGAUGGGUAAUCUUAGGUAUCUC--------- -((((.((.((((..((.(((((((.(((..((((((..((((((...)).-))))..)-)))))..))).))))))).))..)))).)).)).))......--------- ( -28.70, z-score = -2.31, R) >droAna3.scaffold_13266 18214132 95 + 19884421 -UCAAGAUGACUUUACAAGAAUAGCAUUGUCUCGGA------UUUAUAUUU-AAAUACCAUCAGUCACAACGCUAUUCGAGAUGAGUCAUUUUGGACGUAUCC-------- -((((((((((((.....(((((((.((((((.(((------((((....)-)))).))...))..)))).))))))).....))))))))))))........-------- ( -28.80, z-score = -4.80, R) >dp4.chr3 2308612 98 - 19779522 -CGCAAACAACCCAACUGGAAUAGCACUGCCGCUGAGAAAAUUACAAAUGC-AGUCA---UCAGUCACAACGCUAUUCGAGACGGGUUGUUUCAGUCGUACUC-------- -.(.(((((((((..((.(((((((..((..(((((((..((.....))..-..)).---)))))..))..))))))).))..))))))))))..........-------- ( -31.50, z-score = -3.77, R) >droPer1.super_2 2480070 98 - 9036312 -CGCAAACAACCCAACUGGAAUAGCACUGCCGCUGAGAAAAUUACAAAUGC-AGUCA---UCAGUCACAACGCUAUUCGAGACGGGUUGUUUCAGUCGUACUC-------- -.(.(((((((((..((.(((((((..((..(((((((..((.....))..-..)).---)))))..))..))))))).))..))))))))))..........-------- ( -31.50, z-score = -3.77, R) >droWil1.scaffold_180700 3469396 104 + 6630534 CCACAAAUGCCCCACCUCGUAUAGCAUAGUGGCUGACUCUCUCAUAU-CAC-AAUCA---UCAGUUACAACGCUAUUCGAGAUUGGGCAUUUAAGUCAUCGAACUGUUC-- ....((((((((...((((.(((((...((((((((...........-...-.....---))))))))...))))).))))...)))))))).................-- ( -30.91, z-score = -4.52, R) >droVir3.scaffold_12875 9403394 104 - 20611582 -CUCAAGUGCCUUAACCCGAAUGGCAUUUCAACUGAUGUUAUUCCAGAAU--UGUCA---UCAGCCAUAAUGCUAUUCGAGAGAAGACAAUU-AGCCAUUUAAGUGAACCA -......((.(((..(.(((((((((((....((((((..(((....)))--...))---))))....))))))))))).)..))).))...-.................. ( -24.70, z-score = -2.66, R) >droMoj3.scaffold_6496 9158927 106 + 26866924 -CGUAAAUGCCUUGACCCGAAUGGCAUUCCAACUGAUGAUUUUGCAGAAUUGUGUCA---UCGGCCAUAAUGCAAUUCGAGAGAAGGCAAUU-AGCCAGUUAAAUGAGCCA -..(((.((((((..(.(((((.(((((....((((((((...((......))))))---))))....))))).))))).)..)))))).))-)((((......)).)).. ( -30.50, z-score = -2.35, R) >droGri2.scaffold_15245 2671305 91 - 18325388 ----AAGUGCCUUAACUCGAAUGGCAUUUUAACUGAUGAAUUUCUAAAU---UUUCA---UCAGUCACAAUGCAAUUCGAGAAAAGGCAAUA-AGCCAUUCA--------- ----...((((((..(((((((.(((((.(.(((((((((.........---.))))---))))).).))))).)))))))..))))))...-.........--------- ( -34.40, z-score = -7.32, R) >triCas2.ChLG10 4636082 94 + 8806720 -UUUAUUUAACACAACUUCAAUAUCUUCG-GGCUCAGUCACAACAAUGUAC-AAUU----GGCGCCCCAGCCAAAUUUUCCAGGGCGAUUUGUAUUUUUUU---------- -..........((((..((.....(....-)((((.((.(((....)))))-..((----(((......)))))........)))))).))))........---------- ( -17.00, z-score = -1.29, R) >consensus _CCAAAAUAACCUAACUGGAAUAGCAUUGCCGCUGAGUAGAUUCGAUACGC_AGUUA___UCAGUCACAACGCUAUUCGAGAUGGGUAAUUUCAGGCGUGUCC________ .........((((.....((((.((......(((((........................)))))......)).)))).....))))........................ ( -7.34 = -6.98 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:03 2011