
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,266,539 – 4,266,633 |
| Length | 94 |
| Max. P | 0.850382 |

| Location | 4,266,539 – 4,266,633 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.94 |
| Shannon entropy | 0.41517 |
| G+C content | 0.43426 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.66 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4266539 94 - 21146708 -----------------------GAGUGUGUUUUCUUUCGGGUGGGCAUCUGCAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU -----------------------..(((((((...(((((...(((((..(((((((......((((((.....)))))).))))))).)))))...))))).....)))))))... ( -28.00, z-score = -1.71, R) >droSim1.chr2R 2902873 94 - 19596830 -----------------------GAGUGUGUUUUCUUUCGGGUGGGCAUCUGCAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU -----------------------..(((((((...(((((...(((((..(((((((......((((((.....)))))).))))))).)))))...))))).....)))))))... ( -28.00, z-score = -1.71, R) >droSec1.super_1 1912304 94 - 14215200 -----------------------GAGUGUGUUUUCUUUCGGGUGGGCAUCUGCAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU -----------------------..(((((((...(((((...(((((..(((((((......((((((.....)))))).))))))).)))))...))))).....)))))))... ( -28.00, z-score = -1.71, R) >droYak2.chr2L 16922964 94 - 22324452 -----------------------GAGUGUGUUUUCUUUCGGGUGGGCAUCUGCAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCCUUGGCGAGAUUUGUAGCAUACAUU -----------------------..(((((((...(((((...(((((..(((((((......((((((.....)))))).))))))).)))))...))))).....)))))))... ( -30.80, z-score = -2.37, R) >droEre2.scaffold_4929 8275082 94 + 26641161 -----------------------GAGUGUGUUUUCUUUCGGGUGGGCAUCUGCAUUUAUGCAAGUAGCUUAAAUGGCUACGAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU -----------------------..(((((((...(((((...(((((..(((((((......((((((.....)))))).))))))).)))))...))))).....)))))))... ( -30.20, z-score = -2.36, R) >droAna3.scaffold_13266 18155188 96 + 19884421 --------------------CUAGUGUGUU-UUCUUUUGGGGCGGCCAUCUACAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU --------------------...(((((((-.(((((..(((((((......((((((.((.....)).))))))((........)))))))))..).)))).....)))))))... ( -27.80, z-score = -1.60, R) >dp4.chr3 2244790 92 - 19779522 -----------------------GAGUGUG-UUUCUUUUGGGUGUGCGCCUACA-UUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU -----------------------..(((((-((.....(..((.(.((((....-..(((((.((((((.....))))))....)))))......))))).))..).)))))))... ( -27.40, z-score = -1.34, R) >droWil1.scaffold_180701 696980 77 - 3904529 ---------------------------------UCUUUCA-------CUUUGCAUUUAUGCAAGUAGCUUAAAUGGCUACGAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU ---------------------------------.......-------.....((((((.((.....)).))))))((((((((...((((.....))))...))))))))....... ( -19.20, z-score = -1.08, R) >droGri2.scaffold_15245 2607891 80 - 18325388 -----------------------------------UGGCCGCUG--UAGCACCAUUUAUGCAAGUAGCUUAAAUGGCUACAGAAUGCGUUGCUUCGGCGAGAUUUGUUGCAUACAUU -----------------------------------..((..(((--((((..((((((.((.....)).)))))))))))))...)).((((....))))................. ( -24.60, z-score = -1.11, R) >droVir3.scaffold_12875 9310359 105 - 20611582 ----------CGGCUACCGGGUGACGGGUGGAGCCUGCCGACUG--CAGCUACAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUCGGCGAGAUUUGUUGCAUACAUU ----------.(((((((.(....).)))..))))((((((..(--((((..((((((.((.....)).))))))((........))))))).)))))).................. ( -37.70, z-score = -2.05, R) >droMoj3.scaffold_6496 9046404 115 + 26866924 CGGCAGCAGGAGGCGGGCAGCAGGAGGCAGGAGGCAGUCGGCUG--CAGCUACAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUCGGCGAGAUUUGUUGCAUACAUU ..((((((((.(((..(((((.((..((.....))..)).))))--).))).......((((.((((((.....))))))....))))((((....))))..))))))))....... ( -41.60, z-score = -2.27, R) >consensus _______________________GAGUGUGUUUUCUUUCGGGUGGGCAUCUGCAUUUAUGCAAGUAGCUUAAAUGGCUACAAAAUGCGUUGCUUUGGCGAGAUUUGUAGCAUACAUU .......................................((((....)))).....(((((..((((((.....))))))........((((....))))........))))).... (-17.65 = -17.66 + 0.01)

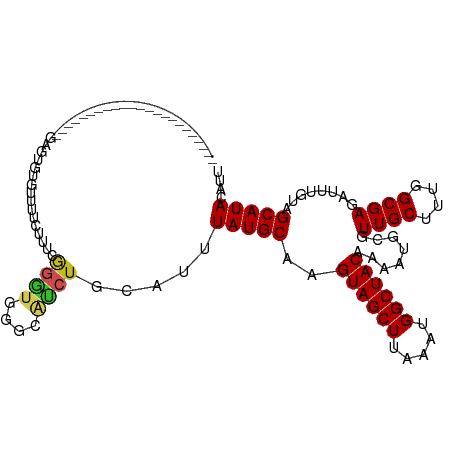

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:57 2011