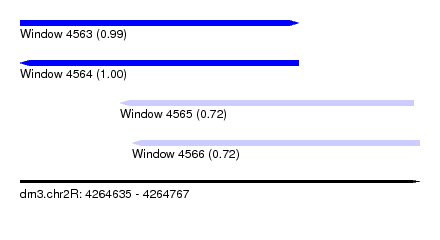
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,264,635 – 4,264,767 |
| Length | 132 |
| Max. P | 0.999716 |

| Location | 4,264,635 – 4,264,727 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Shannon entropy | 0.41155 |
| G+C content | 0.51046 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
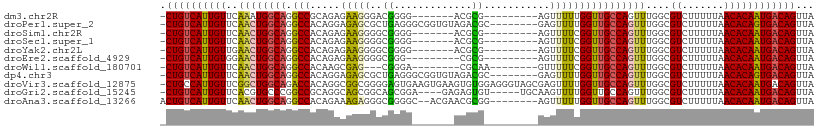
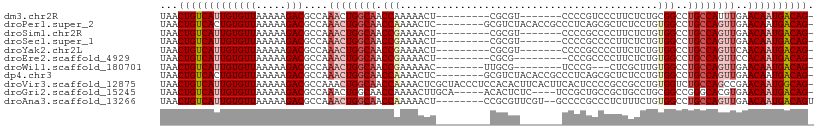
>dm3.chr2R 4264635 92 + 21146708 -CUGUCAUUGUUCAAAUGGCAGGCCGCAGAGAAGGGACGGGG-------ACGCG---------AGUUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -((((((((((......((....)).....((((((((((((-------((...---------.))))))....(((.....)))))))))))...))))))))))... ( -32.00, z-score = -2.83, R) >droPer1.super_2 2415086 100 + 9036312 -CUGUCAUUGUUCAACUGGCAGGCCACAGGAGAGCGCUGAGGGCGGUGUAGACGC--------GAGUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAGUGACAGUUA -((((((((((.....(((....)))..(((((((((..(.(((..(..((((..--------..))))..)..)))...)..))).))))))...))))))))))... ( -37.50, z-score = -2.35, R) >droSim1.chr2R 2900958 92 + 19596830 -CUGUCAUUGUUCAACUGGCAGGCCACAGAGAAGGGGCGGGG-------ACGCG---------AGUUUUCGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -((((((((((((((((((((((((.(......).)))(..(-------((...---------.)))..)..))))))).)))).((.......))))))))))))... ( -36.40, z-score = -3.96, R) >droSec1.super_1 1910391 92 + 14215200 -CUGUCAUUGUUCAACUGGCAGGCCACAGAGAAGGGGCGGGG-------ACGCG---------AGUUUUCGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -((((((((((((((((((((((((.(......).)))(..(-------((...---------.)))..)..))))))).)))).((.......))))))))))))... ( -36.40, z-score = -3.96, R) >droYak2.chr2L 16921017 92 + 22324452 -CUGUCAUUGUUGAACUGGCAGGCCACAGAGAAGGGGCGGGG-------ACGCG---------AGUUUUCGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -((((((((((..((((((((((((.(......).)))(..(-------((...---------.)))..)..)))))))))..))((.......))..))))))))... ( -39.80, z-score = -4.91, R) >droEre2.scaffold_4929 8273196 90 - 26641161 -CUGUCAUUGUGGAACUGGCAGGCCACAGAGAAGGGGCGGG---------CGCG---------AGUUUUCGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -((((((((((((((((((((((((.(......).)))(((---------(...---------.))))....))))))))))(....).......)))))))))))... ( -35.00, z-score = -3.19, R) >droWil1.scaffold_180701 693693 89 + 3904529 -CUGUCAUUGUUCAACUGGCAGGCCACAAGCGAG---CGGGA--------CGCAA--------GUUUUUCGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -(((((((((((((((((((((.((..((((..(---((...--------)))..--------))))...))))))))).)))).((.......))))))))))))... ( -31.10, z-score = -2.88, R) >dp4.chr3 2242660 100 + 19779522 -CUGUCAUUGUUCAACUGGCAGGCCACAGGAGAGCGCUGAGGGCGGUGUAGACGC--------GAGUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAGUGACAGUUA -((((((((((.....(((....)))..(((((((((..(.(((..(..((((..--------..))))..)..)))...)..))).))))))...))))))))))... ( -37.50, z-score = -2.35, R) >droVir3.scaffold_12875 11305919 108 - 20611582 -CUGCCAUUGUUCGGCUGGCAGACCACAGGCGGCGGGGAGUGAAGUGAAGUGUGGAGGGUAGCGAGUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -(((.((((((((.((((.(........).)))).))..((.....(((((((.((.((((((........))))))...)).))).))))...)))))))).)))... ( -29.80, z-score = 0.82, R) >droGri2.scaffold_15245 15721615 99 - 18325388 -CUGUCAUUGUUCACGUGCCCGGCCGCAGGCAGCGGCAGCGGA----GAGAGUGU-----UGCAAGUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA -((((((((((.(((...(((((((((.....)))))..))).----)...))).-----.....(((..((.((((.....)))).))....)))))))))))))... ( -36.40, z-score = -1.95, R) >droAna3.scaffold_13266 18153358 99 - 19884421 ACUGUCAUUGUUCAACUGGCAGGCCACAGAAAGAGGGCGGGGC--ACGAACGCGG--------AGUUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA ((((((((((((((((((((((.(((.(((..(.(..((....--.))..).)..--------..))).)))))))))).)))).((.......))))))))))))).. ( -33.50, z-score = -2.28, R) >consensus _CUGUCAUUGUUCAACUGGCAGGCCACAGAGAAGGGGCGGGG_______ACGCG_________AGUUUUUGGUUGCCAGUUUGGCGUCUUUUUAACACAAUGACAGUUA .((((((((((..((((((((.(((.............................................)))))))))))....((.......))))))))))))... (-21.43 = -21.59 + 0.16)
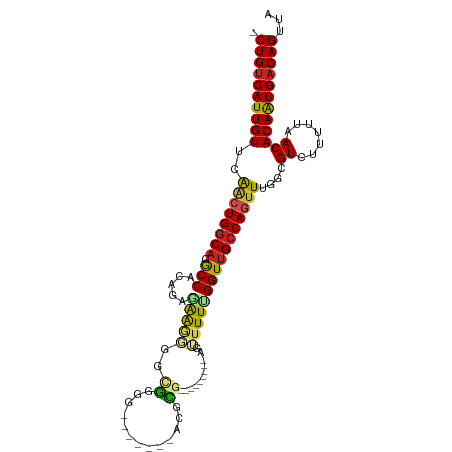
| Location | 4,264,635 – 4,264,727 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Shannon entropy | 0.41155 |
| G+C content | 0.51046 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -20.93 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4264635 92 - 21146708 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACU---------CGCGU-------CCCCGUCCCUUCUCUGCGGCCUGCCAUUUGAACAAUGACAG- ...((((((((((.(((((.....(((.....)))...........---------.(((.-------..((((.........))))..)))..)))))))))))))))- ( -24.50, z-score = -3.59, R) >droPer1.super_2 2415086 100 - 9036312 UAACUGUCACUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACUC--------GCGUCUACACCGCCCUCAGCGCUCUCCUGUGGCCUGCCAGUUGAACAAUGACAG- ...((((((..(((((.....)))))..((((((((.(((......--------((((..............)))).......)))..))))))))......))))))- ( -28.46, z-score = -2.85, R) >droSim1.chr2R 2900958 92 - 19596830 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU---------CGCGU-------CCCCGCCCCUUCUCUGUGGCCUGCCAGUUGAACAAUGACAG- ...(((((((((((((.....)))....(((((((((((((....)---------)).))-------....(((.(......).))).))))))))..))))))))))- ( -31.30, z-score = -5.34, R) >droSec1.super_1 1910391 92 - 14215200 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU---------CGCGU-------CCCCGCCCCUUCUCUGUGGCCUGCCAGUUGAACAAUGACAG- ...(((((((((((((.....)))....(((((((((((((....)---------)).))-------....(((.(......).))).))))))))..))))))))))- ( -31.30, z-score = -5.34, R) >droYak2.chr2L 16921017 92 - 22324452 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU---------CGCGU-------CCCCGCCCCUUCUCUGUGGCCUGCCAGUUCAACAAUGACAG- ...(((((((((((((.....)))....(((((((((((((....)---------)).))-------....(((.(......).))).))))))))..))))))))))- ( -30.70, z-score = -5.34, R) >droEre2.scaffold_4929 8273196 90 + 26641161 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU---------CGCG---------CCCGCCCCUUCUCUGUGGCCUGCCAGUUCCACAAUGACAG- ...(((((((((((.......(....).((((((((..(((....)---------))..---------...(((.(......).))).)))))))).)))))))))))- ( -30.00, z-score = -4.94, R) >droWil1.scaffold_180701 693693 89 - 3904529 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAAAC--------UUGCG--------UCCCG---CUCGCUUGUGGCCUGCCAGUUGAACAAUGACAG- ...(((((((((((((.....)))....((((((((.((.....((--------..(((--------.....---..)))..))))..))))))))..))))))))))- ( -30.60, z-score = -4.23, R) >dp4.chr3 2242660 100 - 19779522 UAACUGUCACUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACUC--------GCGUCUACACCGCCCUCAGCGCUCUCCUGUGGCCUGCCAGUUGAACAAUGACAG- ...((((((..(((((.....)))))..((((((((.(((......--------((((..............)))).......)))..))))))))......))))))- ( -28.46, z-score = -2.85, R) >droVir3.scaffold_12875 11305919 108 + 20611582 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACUCGCUACCCUCCACACUUCACUUCACUCCCCGCCGCCUGUGGUCUGCCAGCCGAACAAUGGCAG- ...(((((((((((((.....)))......((((((((((......((...........................))......)))).))))))....))))))))))- ( -26.63, z-score = -2.49, R) >droGri2.scaffold_15245 15721615 99 + 18325388 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACUUGCA-----ACACUCUC----UCCGCUGCCGCUGCCUGCGGCCGGGCACGUGAACAAUGACAG- ...((((((((((...........(((.....)))..............-----.(((....----((((..(((((.....)))))))))...))).))))))))))- ( -34.30, z-score = -3.72, R) >droAna3.scaffold_13266 18153358 99 + 19884421 UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACU--------CCGCGUUCGU--GCCCCGCCCUCUUUCUGUGGCCUGCCAGUUGAACAAUGACAGU ..((((((((((((((.....)))....((((((((..........--------.(((....))--)....((((.......).))).))))))))..))))))))))) ( -32.00, z-score = -4.30, R) >consensus UAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACU_________CGCGU_______CCCCGCCCCUUCUCUGUGGCCUGCCAGUUGAACAAUGACAG_ ...(((((((((((((.....)))....((((((((.(((...........................................)))..))))))))..)))))))))). (-20.93 = -21.68 + 0.75)
| Location | 4,264,668 – 4,264,765 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Shannon entropy | 0.37091 |
| G+C content | 0.45584 |
| Mean single sequence MFE | -14.89 |
| Consensus MFE | -8.88 |
| Energy contribution | -9.11 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
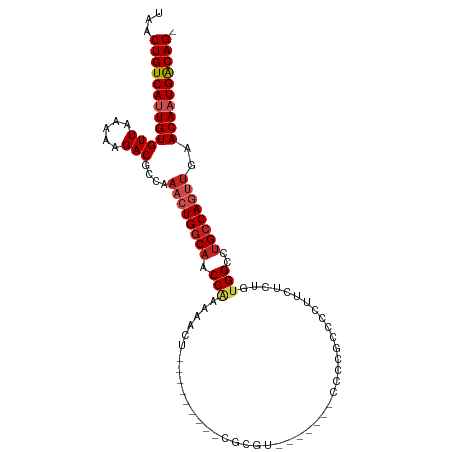
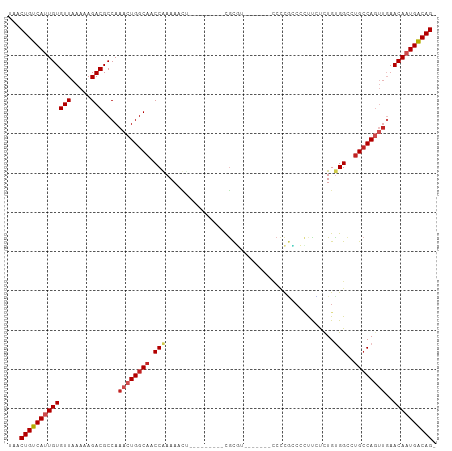
>dm3.chr2R 4264668 97 - 21146708 UAUCCAUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC-AAAAACUCGCGUCCCCGUCC-------- ........((((......-.......((((((((.............))))))))........(((.....)))....-........))))........-------- ( -15.42, z-score = -2.31, R) >droPer1.super_2 2415125 99 - 9036312 UAUCUAUGGCCCUAUCCA-UAUCCCUUCUGCAAUCUAAUUAACUGUCACUGUGUUAAAAAGACGCCAAACUGGCAACC--AAAACUCGCGUCUACACCGCCC----- ....(((((......)))-))........((.......(((((.........)))))..((((((.....(((...))--)......)))))).....))..----- ( -15.10, z-score = -1.81, R) >droSim1.chr2R 2900991 97 - 19596830 UAUCCAUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC-GAAAACUCGCGUCCCCGCCC-------- ........(((.......-.......((((((((.............)))))))).....((((((.....)))...(-((....))).)))..)))..-------- ( -16.72, z-score = -2.29, R) >droSec1.super_1 1910424 97 - 14215200 UAUCCAUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC-GAAAACUCGCGUCCCCGCCC-------- ........(((.......-.......((((((((.............)))))))).....((((((.....)))...(-((....))).)))..)))..-------- ( -16.72, z-score = -2.29, R) >droYak2.chr2L 16921050 97 - 22324452 UAUCCUUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC-GAAAACUCGCGUCCCCGCCC-------- ........(((.......-.......((((((((.............)))))))).....((((((.....)))...(-((....))).)))..)))..-------- ( -16.72, z-score = -2.35, R) >droEre2.scaffold_4929 8273229 95 + 26641161 UAUCCUUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC-GAAAACUCGCG--CCCGCCC-------- ........((((......-.......((((((((.............))))))))........(((.....)))....-........)))--)......-------- ( -17.52, z-score = -2.40, R) >droWil1.scaffold_180701 693726 92 - 3904529 UAUCCCUUUUGCCAUCCUCUAUCCU---UACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAAACUUGCGUCCCG------------ ..........((.............---.....(((..(((((.........)))))..))).(((.....))).............))......------------ ( -9.10, z-score = -0.54, R) >dp4.chr3 2242699 99 - 19779522 UAUCUAUGGCCCUAUCCA-UAUCCCUUCUGCAAUCUAAUUAACUGUCACUGUGUUAAAAAGACGCCAAACUGGCAACC--AAAACUCGCGUCUACACCGCCC----- ....(((((......)))-))........((.......(((((.........)))))..((((((.....(((...))--)......)))))).....))..----- ( -15.10, z-score = -1.81, R) >droGri2.scaffold_15245 15721648 95 + 18325388 UAUCCUUUUACCUAGCCU----------UACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC--AAAACUUGCAACACUCUCUCCGCUGCC ............((((..----------.....(((..(((((.........)))))..))).(((.....)))....--.....................)))).. ( -10.90, z-score = -0.52, R) >droAna3.scaffold_13266 18153392 103 + 19884421 UAUCCUUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACUCCGCGUUCGUGCCCCGCCC--- ........((((......-.......((((((((.............))))))))........(((.....)))...................)))).......--- ( -15.62, z-score = -1.31, R) >consensus UAUCCAUCGCGCUAUCCU_UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACC_GAAAACUCGCGUCCCCGCCC________ .......(((.......................(((..(((((.........)))))..))).(((.....))).............)))................. ( -8.88 = -9.11 + 0.23)

| Location | 4,264,672 – 4,264,767 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.37274 |
| G+C content | 0.43908 |
| Mean single sequence MFE | -14.08 |
| Consensus MFE | -8.60 |
| Energy contribution | -8.55 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
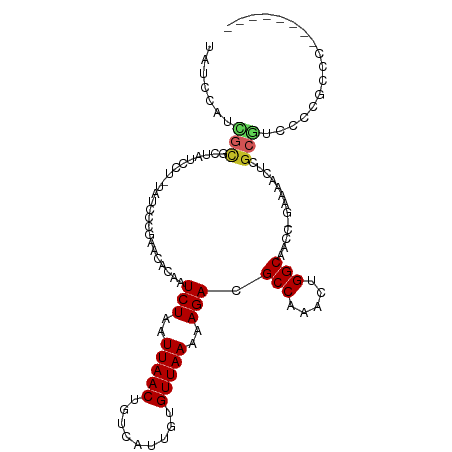
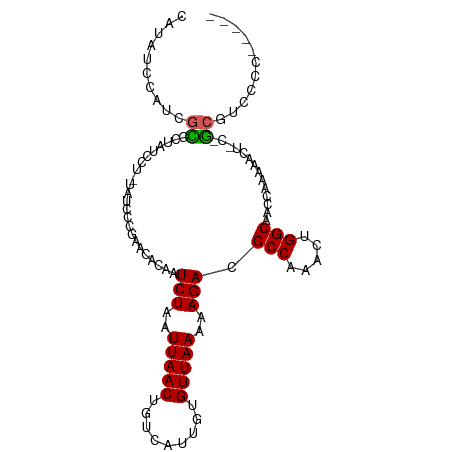
>dm3.chr2R 4264672 95 - 21146708 CAUAUCCAUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACU-C-GCGUCCCC----- ..........((((......-.......((((((((.............))))))))........(((.....)))...........-.-))))....----- ( -15.42, z-score = -2.58, R) >droPer1.super_2 2415129 97 - 9036312 CCUAUCUAUGGCCCUAUCCA-UAUCCCUUCUGCAAUCUAAUUAACUGUCACUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACU--C-GCGUCUACACC-- ......(((((......)))-)).............................((((.....((((((.....(((...))).....--.-)))))))))).-- ( -13.80, z-score = -1.68, R) >droSim1.chr2R 2900995 95 - 19596830 CAUAUCCAUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU-C-GCGUCCCC----- ..........((((......-.......((((((((.............))))))))........(((.....)))...........-.-))))....----- ( -15.42, z-score = -2.00, R) >droSec1.super_1 1910428 95 - 14215200 CAUAUCCAUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU-C-GCGUCCCC----- ..........((((......-.......((((((((.............))))))))........(((.....)))...........-.-))))....----- ( -15.42, z-score = -2.00, R) >droYak2.chr2L 16921054 95 - 22324452 CAUAUCCUUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU-C-GCGUCCCC----- ..........((((......-.......((((((((.............))))))))........(((.....)))...........-.-))))....----- ( -15.42, z-score = -2.05, R) >droEre2.scaffold_4929 8273233 93 + 26641161 CAUAUCCUUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAACU-C-GCGCCC------- ..........((((......-.......((((((((.............))))))))........(((.....)))...........-.-))))..------- ( -17.52, z-score = -2.80, R) >droWil1.scaffold_180701 693727 93 - 3904529 CCUAUCCCUUUUGCCAUCCUCUAUCCU---UACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCGAAAAACUU-GCGUCCC------ ............((.............---.....(((..(((((.........)))))..))).(((.....))).............-)).....------ ( -9.10, z-score = -0.54, R) >dp4.chr3 2242703 95 - 19779522 --UAUCUAUGGCCCUAUCCA-UAUCCCUUCUGCAAUCUAAUUAACUGUCACUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACU--C-GCGUCUACACC-- --....(((((......)))-)).............................((((.....((((((.....(((...))).....--.-)))))))))).-- ( -13.80, z-score = -1.58, R) >droGri2.scaffold_15245 15721652 95 + 18325388 --CAUAUAUCCUUUUACC---UAGCCU---UACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAACUUGCAACACUCUCUCCGC --................---..((..---.....(((..(((((.........)))))..))).(((.....)))...........)).............. ( -9.30, z-score = -0.29, R) >droAna3.scaffold_13266 18153396 101 + 19884421 CAUAUCCUUCGCGCUAUCCU-UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACUCC-GCGUUCGUGCCCC ..........((((......-.......((((((((.............))))))))........(((.....))).............-......))))... ( -15.62, z-score = -1.67, R) >consensus CAUAUCCAUCGCGCUAUCCU_UAUCCCGAACACAAUCUAAUUAACUGUCAUUGUGUUAAAAAGACGCCAAACUGGCAACCAAAAACU_C_GCGUCCCC_____ ..........((.......................(((..(((((.........)))))..))).(((.....)))..............))........... ( -8.60 = -8.55 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:56 2011