| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,258,595 – 4,258,686 |
| Length | 91 |
| Max. P | 0.893278 |

| Location | 4,258,595 – 4,258,686 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.54080 |
| G+C content | 0.36417 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -9.52 |
| Energy contribution | -11.25 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
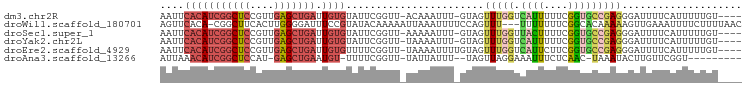
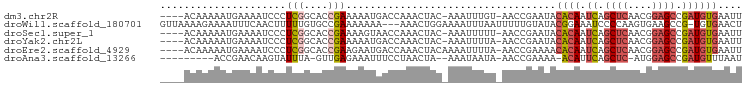
>dm3.chr2R 4258595 91 + 21146708 AAUUCACAUCGGCUCCGUUGAGCUGAUUGUGUAUUCGGUU-ACAAAUUU-GUAGUUUGGUCAUUUUUCGGUGCCGAGGGAUUUUCAUUUUUGU---- (((((...(((((.(((...((((((........))))))-.(((((..-...))))).........))).))))).)))))...........---- ( -24.00, z-score = -2.17, R) >droWil1.scaffold_180701 3216895 93 - 3904529 AGUUCACA-CGGCUUCACUUGGGGAUUUCCGUAUACAAAAAUUAAAUUUUCCAGUUU---UUUUUUUCGGCACAAAAAGUUGAAAUUUUCUUUUAAC .......(-(((((((....))))....)))).........................---....(((((((.......)))))))............ ( -9.60, z-score = 0.95, R) >droSec1.super_1 1904354 91 + 14215200 AAUUCACAUCGGCUCCGUUGAGCUGAUUGUGUAUUCGGUU-AAAAAUUU-GUAGUUUGGUUACUUUUCGGUGCCGAGGGAUUUUCAUUUUUGU---- (((((...(((((.(((...((((((........))))))-........-((((.....))))....))).))))).)))))...........---- ( -22.00, z-score = -1.58, R) >droYak2.chr2L 16915126 91 + 22324452 AAUUCACAUCGGCUCCGUUGAGCUGAUUGUGUAUUCGGUU-UAAAAUUU-GUAGUUUGGUCAUUUUUCGGUGCCGAGGGAUUUUCAUUUUUGU---- (((((...(((((.((((((((((((........))))))-))).....-...(......)......))).))))).)))))...........---- ( -23.00, z-score = -2.07, R) >droEre2.scaffold_4929 8267405 92 - 26641161 AAUUCACAUCGGCUCCGUUGAGCUGAUUGUGUUUUCGGUU-UAAAAUUUUGUAGUUUGGUCAUUCUUCGGUGCCGAGGGAUUUUCAUUUUUGU---- ....(((((((((((....))))))).)))).........-...............(((..((((((((....))))))))..))).......---- ( -24.30, z-score = -2.45, R) >droAna3.scaffold_13266 18147728 82 - 19884421 AUUAAACAUCGGCUCCAU-GAGCUGAAUGU-UUUUCGGUU-UAUUAUUU--UAGUUAGGAAAUUUCUCAAC-UAAAUACUUGUUCGGU--------- ...((((((((((((...-)))))).))))-)).((((..-......((--(((((..((....))..)))-)))).......)))).--------- ( -19.66, z-score = -3.14, R) >consensus AAUUCACAUCGGCUCCGUUGAGCUGAUUGUGUAUUCGGUU_UAAAAUUU_GUAGUUUGGUCAUUUUUCGGUGCCGAGGGAUUUUCAUUUUUGU____ ....(((((((((((....))))))).))))................................((((((....)))))).................. ( -9.52 = -11.25 + 1.73)
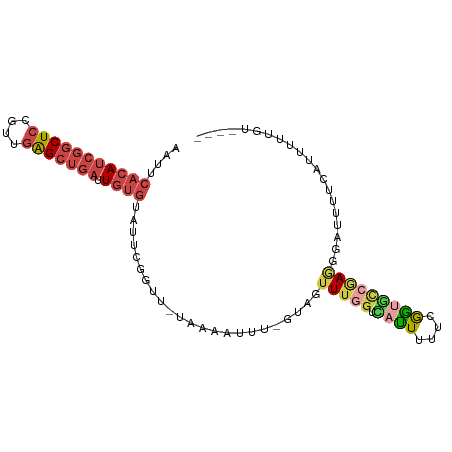
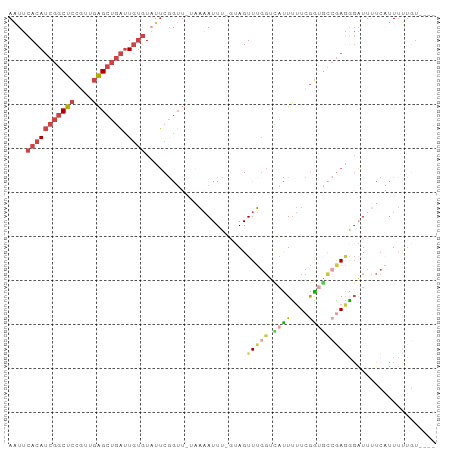
| Location | 4,258,595 – 4,258,686 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.54080 |
| G+C content | 0.36417 |
| Mean single sequence MFE | -13.81 |
| Consensus MFE | -6.47 |
| Energy contribution | -7.30 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
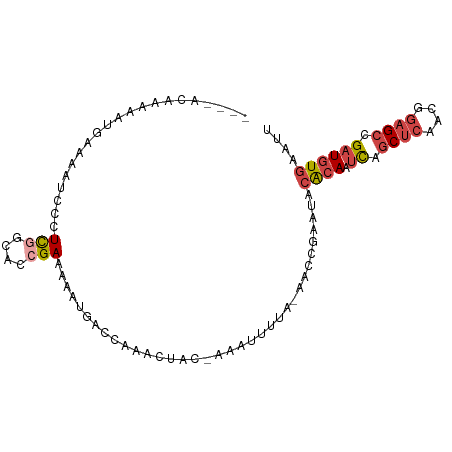
>dm3.chr2R 4258595 91 - 21146708 ----ACAAAAAUGAAAAUCCCUCGGCACCGAAAAAUGACCAAACUAC-AAAUUUGU-AACCGAAUACACAAUCAGCUCAACGGAGCCGAUGUGAAUU ----.............(((.(((((.(((..............(((-(....)))-)...((........)).......))).))))).).))... ( -15.00, z-score = -1.77, R) >droWil1.scaffold_180701 3216895 93 + 3904529 GUUAAAAGAAAAUUUCAACUUUUUGUGCCGAAAAAAA---AAACUGGAAAAUUUAAUUUUUGUAUACGGAAAUCCCCAAGUGAAGCCG-UGUGAACU (((.........(((((..(((((..........)))---))..))))).............(((((((...((.......))..)))-))))))). ( -9.90, z-score = 0.97, R) >droSec1.super_1 1904354 91 - 14215200 ----ACAAAAAUGAAAAUCCCUCGGCACCGAAAAGUAACCAAACUAC-AAAUUUUU-AACCGAAUACACAAUCAGCUCAACGGAGCCGAUGUGAAUU ----.......(((((((...(((....)))...(((.......)))-..))))))-)........((((.((.((((....)))).)))))).... ( -15.50, z-score = -2.28, R) >droYak2.chr2L 16915126 91 - 22324452 ----ACAAAAAUGAAAAUCCCUCGGCACCGAAAAAUGACCAAACUAC-AAAUUUUA-AACCGAAUACACAAUCAGCUCAACGGAGCCGAUGUGAAUU ----.................(((....)))................-........-.........((((.((.((((....)))).)))))).... ( -13.50, z-score = -1.71, R) >droEre2.scaffold_4929 8267405 92 + 26641161 ----ACAAAAAUGAAAAUCCCUCGGCACCGAAGAAUGACCAAACUACAAAAUUUUA-AACCGAAAACACAAUCAGCUCAACGGAGCCGAUGUGAAUU ----.............(((.(((((.(((..((.(((..................-..............)))..))..))).))))).).))... ( -14.85, z-score = -1.99, R) >droAna3.scaffold_13266 18147728 82 + 19884421 ---------ACCGAACAAGUAUUUA-GUUGAGAAAUUUCCUAACUA--AAAUAAUA-AACCGAAAA-ACAUUCAGCUC-AUGGAGCCGAUGUUUAAU ---------............((((-((((.((....)).))))))--))......-.......((-(((((..((((-...)))).)))))))... ( -14.10, z-score = -2.38, R) >consensus ____ACAAAAAUGAAAAUCCCUCGGCACCGAAAAAUGACCAAACUAC_AAAUUUUA_AACCGAAUACACAAUCAGCUCAACGGAGCCGAUGUGAAUU .....................(((....)))...................................((((.((.((((....)))).)))))).... ( -6.47 = -7.30 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:53 2011