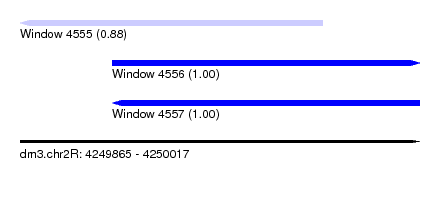
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,249,865 – 4,250,017 |
| Length | 152 |
| Max. P | 0.999910 |

| Location | 4,249,865 – 4,249,980 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Shannon entropy | 0.43810 |
| G+C content | 0.43276 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.882721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4249865 115 - 21146708 CUCAUACGCAGACCAAUUACUCCACGCUCAUUUUAGCGUAGUGAUUGGGUUGCACUUGCGAUUGGCAUCAGGGGUCCAAG-AUUCUCCACAAUUAGCCGUAUUAAAGAUGUAAUGU ....(((((((.(((((((((..(((((......)))))))))))))).)))).(((.....((((....((((......-...)))).......)))).....)))..))).... ( -31.60, z-score = -1.60, R) >droSim1.chr2R_random 1390107 115 - 2996586 CUCAUACGCAGAGCAAUUACUGCACGCGCAUUUUAGCGUAGUGAUUGGGUUGCACUUGAGAUUGGCAACAAGGGUCCAAG-AUUCUCCACAAUUCGACGUAUUAAAGAUGUAAUGU ...(((((..((((((((....(((((((......)))).)))....)))))).((((.(((((....))...)))))))-............))..))))).............. ( -28.50, z-score = -0.48, R) >droSec1.super_1 1895875 115 - 14215200 CUCAUACGCAGACCAAUUACUGCACGCGCAUUUUAGCGUAGUGAUUGGGUUGCACUUGAGAUUGGCAACAAGGGUCCAAG-AUUCUCCACAAUUCGACGUAUUAAAGAUGUAAUGU ...((((((((.(((((((((..((((........))))))))))))).)))).((((.(((((....))...)))))))-.................)))).............. ( -30.90, z-score = -1.45, R) >droYak2.chr2L 16905840 108 - 22324452 CUCAUAUGCACCUCAAUAACU------UCACUUUAGCGUUGUGAUUGGGUUGUACUUGCGAUUGGCAACAUGGGUUCAAG-ACUCUCCACAAUUCGACGUAUUAAAGAUGU-AUGU ...(((((((...........------...(((((((((((.(((((((..((.((((....((....))......))))-))...)).)))))))))))..))))).)))-)))) ( -22.26, z-score = 0.27, R) >droEre2.scaffold_4929 8257772 113 + 26641161 CUCAUACGCAGCUCAAUUACUUGACGUGCAUUUUAGCGUUGUGAUUGGGUUGCACUUGCGAUUGGCACCAGGAGUCCGAG-AUUCUCCACAAUUCGACGUAUUAAAGAUGUAAU-- ....((((((((((((((((..(((((........))))))))))))))))))...((((.((((.....((((......-...)))).....))))))))........)))..-- ( -32.80, z-score = -1.83, R) >droAna3.scaffold_13266 18139357 113 + 19884421 CUCAGAUGCAGCUCAGCCACAA-ACGCGAAAACGUGUUUUGUGACUGAGUUGCAGAUGAGAAGGGCAACAGAUAUCCGGGAACGGACCAAUUAUAAUUGUAUUAAAUGUUUCGU-- ((((..((((((((((.(((((-(((((....)))).)))))).))))))))))..))))...((.((((....((((....)))).((((....)))).......))))))..-- ( -43.10, z-score = -5.58, R) >consensus CUCAUACGCAGAUCAAUUACUGCACGCGCAUUUUAGCGUAGUGAUUGGGUUGCACUUGAGAUUGGCAACAGGGGUCCAAG_AUUCUCCACAAUUCGACGUAUUAAAGAUGUAAUGU ...((((((((.(((((((((..((((........))))))))))))).)))).((((.(((((....))...)))))))..................)))).............. (-18.24 = -19.13 + 0.90)

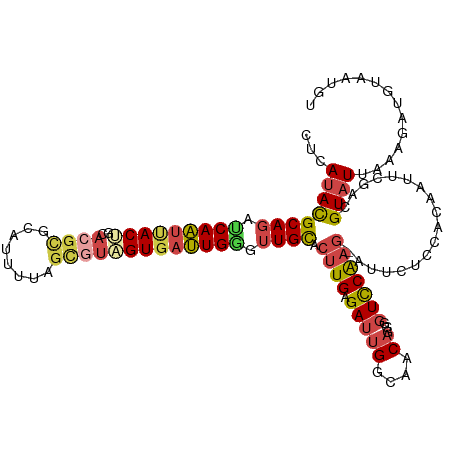
| Location | 4,249,900 – 4,250,017 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.49568 |
| G+C content | 0.46439 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4249900 117 + 21146708 CUUGGACCCCUGAUGCCAAUCGCAAGUGCAACCCAAUCACUACGCUAAAAUGAGCGUGGAGUAAUUGGUCUGCGUAUGAGUUUGGCAUCAGGAUCGAAAACUUUC--AUUUUUUUCUGG- .(..((..(((((((((((.(.((..((((..((((((.(((((((......))))))).)..)))))..))))..)).).)))))))))))...(((....)))--.......))..)- ( -43.50, z-score = -4.68, R) >droSim1.chr2R_random 1390142 120 + 2996586 CUUGGACCCUUGUUGCCAAUCUCAAGUGCAACCCAAUCACUACGCUAAAAUGCGCGUGCAGUAAUUGCUCUGCGUAUGAGUUUGGCAUCAGGAUCGAUAACUAUCCAAUUUUUUUUUUGG ....((.(((.(.((((((.((((..((((...((((.(((((((........))))..))).))))...))))..)))).)))))).)))).)).........((((........)))) ( -34.60, z-score = -2.73, R) >droSec1.super_1 1895910 119 + 14215200 CUUGGACCCUUGUUGCCAAUCUCAAGUGCAACCCAAUCACUACGCUAAAAUGCGCGUGCAGUAAUUGGUCUGCGUAUGAGUUUGGCAUCAGGAUCGAGAACUAUCCAAUUUUUUUUUUG- (((((...((((.((((((.((((..((((..(((((.(((((((........))))..))).)))))..))))..)))).)))))).)))).))))).....................- ( -40.10, z-score = -4.25, R) >droYak2.chr2L 16905874 111 + 22324452 CUUGAACCCAUGUUGCCAAUCGCAAGUACAACCCAAUCACAACGCUAAA------GUGAAGUUAUUGAGGUGCAUAUGAGUUUGGCAUCAGGAUCGAAGACUGUCC-ACUUUGUUCGG-- ..(((((...((.((((((.(.((.((...(((((((.((..(((....------)))..)).)))).)))..)).)).).)))))).))((((........))))-.....))))).-- ( -27.90, z-score = -0.81, R) >droEre2.scaffold_4929 8257805 114 - 26641161 CUCGGACUCCUGGUGCCAAUCGCAAGUGCAACCCAAUCACAACGCUAAAAUGCACGUCAAGUAAUUGAGCUGCGUAUGAGUUUGGCAUCAGGAUCGAAGACUAUCCAUGUUCGG------ ..(((((((((((((((((..((....))..............(((...(((((..((((....))))..)))))...)))))))))))))))..((......))...))))).------ ( -36.40, z-score = -2.42, R) >droAna3.scaffold_13266 18139391 117 - 19884421 CCCGGAUAUCUGUUGCCCUUCUCAUCUGCAACUCAGUCACAAAACACGUUUUCGCGUUU-GUGGCUGAGCUGCAUCUGAGCCUGGCAACAGCACCAAGGAUGAUCCACUAGUGUCCAG-- ...((((((((((((((...((((..((((.((((((((((((.(.((....)).))))-))))))))).))))..))))...))))))))......((.....))....))))))..-- ( -52.00, z-score = -6.44, R) >consensus CUUGGACCCCUGUUGCCAAUCGCAAGUGCAACCCAAUCACAACGCUAAAAUGCGCGUGAAGUAAUUGAGCUGCGUAUGAGUUUGGCAUCAGGAUCGAAAACUAUCCAAUUUUGUUCUG__ .((((...((((.((((((.(.((..((((...((((.(((((((........)))))..)).))))...))))..)).).)))))).)))).))))....................... (-21.86 = -22.83 + 0.98)


| Location | 4,249,900 – 4,250,017 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.49568 |
| G+C content | 0.46439 |
| Mean single sequence MFE | -45.08 |
| Consensus MFE | -29.96 |
| Energy contribution | -29.83 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.86 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.84 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4249900 117 - 21146708 -CCAGAAAAAAAU--GAAAGUUUUCGAUCCUGAUGCCAAACUCAUACGCAGACCAAUUACUCCACGCUCAUUUUAGCGUAGUGAUUGGGUUGCACUUGCGAUUGGCAUCAGGGGUCCAAG -.......(((((--....))))).((((((((((((((.(.((...((((.(((((((((..(((((......)))))))))))))).))))...)).).))))))))).))))).... ( -45.50, z-score = -5.62, R) >droSim1.chr2R_random 1390142 120 - 2996586 CCAAAAAAAAAAUUGGAUAGUUAUCGAUCCUGAUGCCAAACUCAUACGCAGAGCAAUUACUGCACGCGCAUUUUAGCGUAGUGAUUGGGUUGCACUUGAGAUUGGCAACAAGGGUCCAAG ((((........)))).........((((((..((((((.((((...((((..((((((((..((((........))))))))))))..))))...)))).))))))...)))))).... ( -43.70, z-score = -4.98, R) >droSec1.super_1 1895910 119 - 14215200 -CAAAAAAAAAAUUGGAUAGUUCUCGAUCCUGAUGCCAAACUCAUACGCAGACCAAUUACUGCACGCGCAUUUUAGCGUAGUGAUUGGGUUGCACUUGAGAUUGGCAACAAGGGUCCAAG -........................((((((..((((((.((((...((((.(((((((((..((((........))))))))))))).))))...)))).))))))...)))))).... ( -44.30, z-score = -5.32, R) >droYak2.chr2L 16905874 111 - 22324452 --CCGAACAAAGU-GGACAGUCUUCGAUCCUGAUGCCAAACUCAUAUGCACCUCAAUAACUUCAC------UUUAGCGUUGUGAUUGGGUUGUACUUGCGAUUGGCAACAUGGGUUCAAG --..((((...((-(..(((.........))).((((((.(.((..((((.((((((.((..(.(------....).)..)).)))))).))))..)).).)))))).)))..))))... ( -28.40, z-score = -0.69, R) >droEre2.scaffold_4929 8257805 114 + 26641161 ------CCGAACAUGGAUAGUCUUCGAUCCUGAUGCCAAACUCAUACGCAGCUCAAUUACUUGACGUGCAUUUUAGCGUUGUGAUUGGGUUGCACUUGCGAUUGGCACCAGGAGUCCGAG ------.......(((((.(....)..(((((.((((((.(.((...(((((((((((((..(((((........))))))))))))))))))...)).).)))))).)))))))))).. ( -44.10, z-score = -4.25, R) >droAna3.scaffold_13266 18139391 117 + 19884421 --CUGGACACUAGUGGAUCAUCCUUGGUGCUGUUGCCAGGCUCAGAUGCAGCUCAGCCAC-AAACGCGAAAACGUGUUUUGUGACUGAGUUGCAGAUGAGAAGGGCAACAGAUAUCCGGG --(((((((((((.((.....))))))))((((((((...((((..((((((((((.(((-(((((((....)))).)))))).))))))))))..))))...))))))))...))))). ( -64.50, z-score = -8.27, R) >consensus __CAGAAAAAAAUUGGAUAGUCUUCGAUCCUGAUGCCAAACUCAUACGCAGAUCAAUUACUGCACGCGCAUUUUAGCGUAGUGAUUGGGUUGCACUUGAGAUUGGCAACAGGGGUCCAAG .....................(((.(((((((.((((((.(.((...((((.((((((((...((((........)))).)))))))).))))...)).).)))))).)).))))).))) (-29.96 = -29.83 + -0.13)

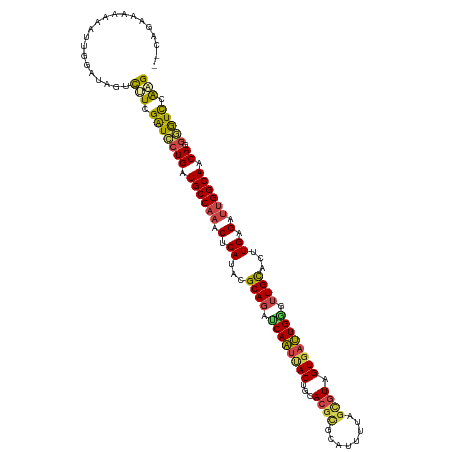
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:49 2011