| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,222,041 – 4,222,134 |
| Length | 93 |
| Max. P | 0.600254 |

| Location | 4,222,041 – 4,222,134 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 59.30 |
| Shannon entropy | 0.77394 |
| G+C content | 0.59660 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -7.84 |
| Energy contribution | -8.05 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
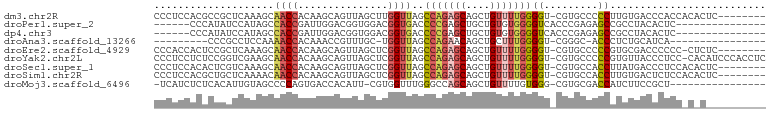
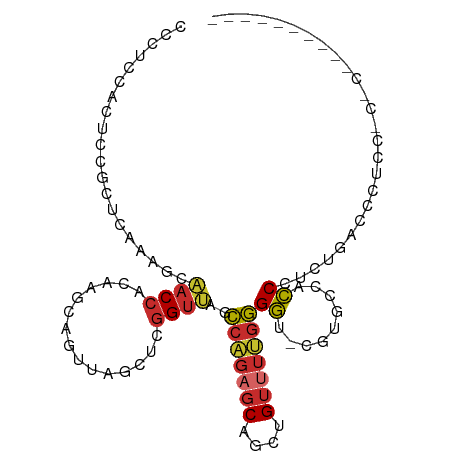
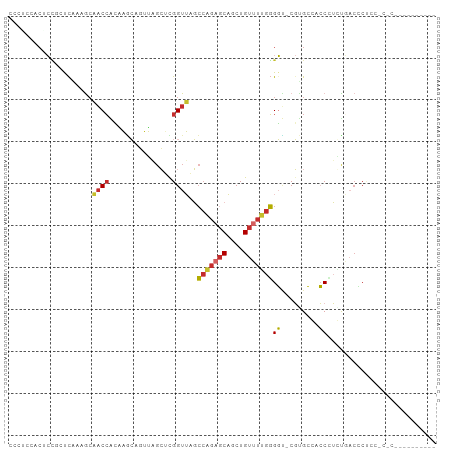
>dm3.chr2R 4222041 93 + 21146708 CCCUCCACGCCGCUCAAAGCAACCACAAGCAGUUAGCUUGGUUAGCCAGAGCAGCUGUUUUGGGGU-CGUGCCCCCUUGUGACCCACCACACUC-------- .....((((((((.....))..(((.((((((((.((((((....))).)))))))))))))))).-))))......((((......))))...-------- ( -30.10, z-score = -1.67, R) >droPer1.super_2 2374564 81 + 9036312 ------CCCAUAUCCAUAGCCACCGAUUGGACGGUGGACGGUGACCCCGAGCUGCUGUGUGGGGUCACCCGAGAGCCGCCUACACUC--------------- ------......((((...........)))).(((((..((((((((((.((....)).))))))))))(....)))))).......--------------- ( -32.40, z-score = -1.76, R) >dp4.chr3 2203196 81 + 19779522 ------CCCAUAUCCAUAGCCACCGAUUGGACGGUGGACGGUGACCCCGAGCUGCUGUGUGGGGUCACCCGAGAGCCGCCUACACUC--------------- ------......((((...........)))).(((((..((((((((((.((....)).))))))))))(....)))))).......--------------- ( -32.40, z-score = -1.76, R) >droAna3.scaffold_13266 18112149 74 - 19884421 ---------CCCGCCUCCAAAACCACAAACCGUUUGC-UGGUUAGCCAGAACAGCUGCUUUGGGGU-CGGGC-ACCCUCUGCAUCA---------------- ---------((((.((((((((((((((.....))).-)))))(((.((.....))))))))))).-)))).-.............---------------- ( -21.30, z-score = -0.04, R) >droEre2.scaffold_4929 8230471 92 - 26641161 CCCACCACUCCGCUCAAAGCAACCACAAGCAGUUAGCUCGGUUAGCCAGAGCAGCUGUUUUGGGGU-CGUGCCCCCGUGCGACCCCCC-CUCUC-------- ...........((.....))......((((((((.((((((....)).)))))))))))).(((((-((..(....)..)))))))..-.....-------- ( -35.30, z-score = -4.12, R) >droYak2.chr2L 16877881 100 + 22324452 CCCUCCUCUCCGGUCGAAGCAACCACAAGCAGUUAGCUCGGUUAGCCAGAGCAGCUGUUUUGGGGU-CGUGCCCCCGUGUUACCCUCC-CACAUCCCACCUC ...........(((...((((.....((((((((.((((((....)).)))))))))))).((((.-....))))..)))))))....-............. ( -26.70, z-score = -0.88, R) >droSec1.super_1 1868206 93 + 14215200 CCCUCCACACUCGUCAAAGCAACCACAAGCAGUUAGCUCGGUUAGCCAGAGCAGCUGUUUUGGGGU-CGUGCCACCUUAUGACCCUCCACACUC-------- .......................((.((((((((.((((((....)).))))))))))))))((((-((((......)))))))).........-------- ( -27.80, z-score = -2.30, R) >droSim1.chr2R 2883668 93 + 19596830 CCCUCCACGCUGCUCAAAACAACCACAAGCAGUUAGCUCGGUUAGCCAGAGCAGCUGUUUUGGGGU-CGUGCCACCUUGUGACUCUCCACACUC-------- (((...(((((((((.....((((...(((.....))).)))).....))))))).))...)))..-..........((((......))))...-------- ( -26.10, z-score = -0.66, R) >droMoj3.scaffold_6496 8984994 83 - 26866924 -UCAUCUCUCACAUUGUAGCCCCAGUGACCACAUU-CGUGGUUUGGGCCAGCAGCUGUUUUGUGGG-CGUGCGACCAUCUUCCGCU---------------- -....(.((((((..(((((((((..((((((...-.))))))))))......)))))..))))))-.).(((.........))).---------------- ( -24.70, z-score = -0.44, R) >consensus CCCUCCACUCCGCUCAAAGCAACCACAAGCAGUUAGCUCGGUUAGCCAGAGCAGCUGUUUUGGGGU_CGUGCCACCCUCUGACCCUCC_C_C__________ ....................((((...............))))..(((((((....)))))))....................................... ( -7.84 = -8.05 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:43 2011