
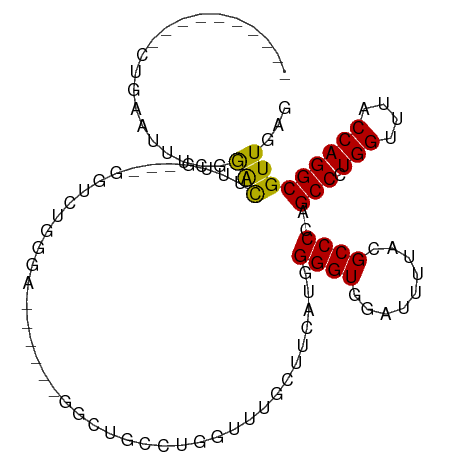
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,221,225 – 4,221,381 |
| Length | 156 |
| Max. P | 0.839268 |

| Location | 4,221,225 – 4,221,316 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.30 |
| Shannon entropy | 0.57781 |
| G+C content | 0.54437 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -13.41 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
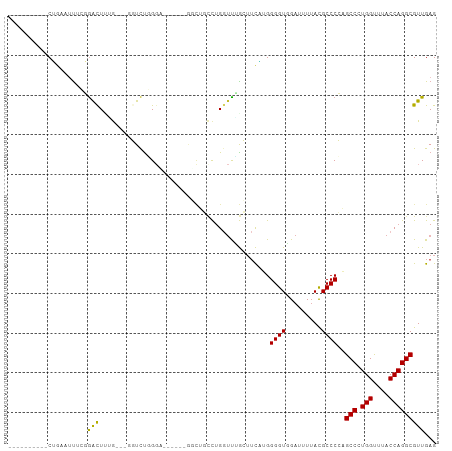
>dm3.chr2R 4221225 91 + 21146708 ----------CUGAAUUUCGGAUUGGG---GGUCUGGU----CUGAGCUGCCUGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAG ----------((.((((((((((..(.---...)..))----)))))..(((((((..(((...(((((((.......))))))).....)))..)))))))))).)) ( -40.30, z-score = -2.74, R) >droSim1.chr2R 2882882 92 + 19596830 ----------CUGAAUUUCGGAUUGUG---GGUCUGGGU---CUGGGCUGCCAGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAG ----------((.(((.((((((....---.))))))((---((((...((((((...(((....((((((.......)))))))))))))))...))))))))).)) ( -38.50, z-score = -1.86, R) >droSec1.super_1 1867438 86 + 14215200 ----------CUGAAUUUCGGAUUGUG---GGUCUGGG---------CUGCCUGUUCUGCUUCAUGGGGUGGACUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAG ----------.((((...((((..(..---(((.....---------..)))..))))).)))).((((((.......))))))((((((((....))))).)))... ( -29.50, z-score = -0.40, R) >droYak2.chr2L 16876994 81 + 22324452 ----------CUGAAUUUCGGGCUUGG----GUUUGG-------------CCUGGGUUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAG ----------.......((((((((((----....((-------------((.((((.......(((((((.......))))))))))).))))..)))))).)))). ( -34.51, z-score = -2.23, R) >droEre2.scaffold_4929 8229657 98 - 26641161 ----------CUGAAAUUCGGGCUUGGUCUGGUCUGGGCUGCCUGGGCUGCCUGGUCCGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGCUUACCAGGCGUUGAG ----------.......(((((((((((..((((.((((((...((((.....(((((((((....)))))))))....)))))))))).)))).))))))).)))). ( -48.00, z-score = -2.71, R) >droAna3.scaffold_13266 18111546 102 - 19884421 UUUAGUCCUGCUCCUGCUCCAGCUCCA---GGUCCUGGAC-CUGGAACUUGCUGCUUGGC--CAAGGGGUGGAUUUAACGCCCCUGCCCUGGUUUACCAGGCGUUGAG .........((....))..((((((((---((((...)))-)))))....))))((..((--..(((((((.......)))))))(((.(((....))))))))..)) ( -42.80, z-score = -2.13, R) >droVir3.scaffold_12875 11359236 87 - 20611582 ---------GCAACUUUAACGACUUCG----CUUU-GGA------GUUACUUUGGUUAAGUUUU-GGGGUUGAUUUUACGCCCGUGCCAUGGCUUACCAGGCGUUGAG ---------.(((((((((.(((((.(----((..-((.------....))..))).)))))))-)))))))..((.(((((.(.((....))....).))))).)). ( -23.30, z-score = -0.70, R) >droMoj3.scaffold_6496 17883927 87 + 26866924 ---------GCGACUUUAACGACUUUG----CUUU-GGA------GUUACUUUGGUUAAGUUUC-GGGGUUGAUUUUACGCCCGUGCCAUGGCUUACCAGGCGUUGAG ---------......((((((((((.(----((..-((.------....))..))).))))).(-(((((.......)).)))).(((.(((....))))))))))). ( -22.60, z-score = -0.35, R) >droGri2.scaffold_15245 15767422 89 - 18325388 ---------GCGACUUUAACGACUUUG----CUUUUGGA------GUUACUUUGGUUAAGUUUUUGGGGUUGAUUUUAUGCCCGUGCCAUGGCUUACCAGGCGUUGAG ---------.(((((((((.(((((.(----((...(..------....)...))).))))).))))))))).........(..((((.(((....)))))))..).. ( -22.60, z-score = -0.78, R) >consensus __________CUGAAUUUCGGACUUUG___GGUCUGGGA______GGCUGCCUGGUUUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAG ....................(((...........................................((((.........))))..(((.(((....)))))))))... (-13.41 = -12.87 + -0.54)

| Location | 4,221,247 – 4,221,354 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.38 |
| Shannon entropy | 0.53699 |
| G+C content | 0.49304 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.21 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4221247 107 + 21146708 ---------GGUCUGAGCUGCCUGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUUGUUGGCUC-- ---------.....((((((((((((..(((...(((((((.......))))))).....)))..)))))))...((.((((.(((((..........))))).)))).)))))))-- ( -44.50, z-score = -3.82, R) >droSim1.chr2R 2882904 108 + 19596830 --------GGGUCUGGGCUGCCAGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUUGUUGGCUC-- --------..((((((...((((((...(((....((((((.......)))))))))))))))...))))))((..(.((((.(((((..........))))).)))).)..))..-- ( -47.40, z-score = -3.82, R) >droSec1.super_1 1867454 108 + 14215200 --------GGGUCUGGGCUGCCUGUUCUGCUUCAUGGGGUGGACUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUUGUUGGCUC-- --------..((((((...(((......(((....((((((.......)))))))))...)))...))))))((..(.((((.(((((..........))))).)))).)..))..-- ( -39.90, z-score = -1.79, R) >droYak2.chr2L 16877015 98 + 22324452 ------------------GGCCUGGGUUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUCUACGGUAUUUGUUGGCUC-- ------------------.((((((...(((...(((((((.......))))))).....)))...))))))((..(.((((.(((((..........))))).)))).)..))..-- ( -41.60, z-score = -3.52, R) >droEre2.scaffold_4929 8229677 116 - 26641161 GGUCUGGGCUGCCUGGGCUGCCUGGUCCGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGCUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUUGUUGGCUC-- ((((..((...))..))))(((((((..(((...(((((((.......))))))).....)))..)))))))((..(.((((.(((((..........))))).)))).)..))..-- ( -52.70, z-score = -3.38, R) >droAna3.scaffold_13266 18111585 101 - 19884421 -------------GGAACUUGCUGCUUGGC--CAAGGGGUGGAUUUAACGCCCCUGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUCUACGGUAUUUGUUGGCUC-- -------------..........((..(((--..(((((((.......))))))))))((((....))))))((..(.((((.(((((..........))))).)))).)..))..-- ( -36.20, z-score = -1.73, R) >dp4.chr3 2202533 107 + 19779522 ----------GUUCCUUUUUUUCUGUUGGGCUCU-GGGGUUGAUUUAACGCCCCGUCCCUGGUCUACCAGGCGUCGAGGAAUUCCGAAAAAGUGUUUUUACGGUAUUUCUGUUGGCAC ----------(((((((..(.((((.((((((.(-(((((.........)))))).....)))))).)))).)..))))))).........(((((...((((.....)))).))))) ( -29.90, z-score = -0.48, R) >droPer1.super_2 2373915 107 + 9036312 ----------GUUCCUUUUUUUCUGUUGGGCUCU-GGGGUUGAUUUAACGCCCCGUCCCUGGUCUACCAGGCGUCGAGGAAUUCCGAAAAAGUGUUUUUACGGUAUUUCUGUUGGCAC ----------(((((((..(.((((.((((((.(-(((((.........)))))).....)))))).)))).)..))))))).........(((((...((((.....)))).))))) ( -29.90, z-score = -0.48, R) >droWil1.scaffold_180701 640680 87 + 3904529 -----------------------AGCUUUUUUUU-GGGGUUGAUUUUACGCCCCGUCCCUGGCUUACCAGGCGUUGAGAAAUUCCGUAAA-GUGUUUUUACGGUAUUUUUGC------ -----------------------(((........-(((((.........)))))...(((((....))))).))).((((((.(((((((-(...)))))))).))))))..------ ( -24.70, z-score = -1.83, R) >droVir3.scaffold_12875 11359258 98 - 20611582 ------------GGAGUUACUUUGGUUAAGUUUU-GGGGUUGAUUUUACGCCCGUGCCAUGGCUUACCAGGCGUUGAGAAAUUCCGUAAA-GUGUUUUUACGGUAUUUGCGC------ ------------((((((((((..(.......).-.))).))))))).((((..((((.(((....)))))))..)..((((.(((((((-(...)))))))).))))))).------ ( -24.10, z-score = 0.04, R) >droMoj3.scaffold_6496 17883949 98 + 26866924 ------------GGAGUUACUUUGGUUAAGUUUC-GGGGUUGAUUUUACGCCCGUGCCAUGGCUUACCAGGCGUUGAGAAAUUCCGUAAA-GUGUUUUUACGGUAUUUGCGC------ ------------((((((.(((...........(-(((((.......)).)))).(((.(((....))))))...))).))))))(((((-.(((....)))...)))))..------ ( -24.80, z-score = -0.04, R) >droGri2.scaffold_15245 15767445 99 - 18325388 ------------GGAGUUACUUUGGUUAAGUUUUUGGGGUUGAUUUUAUGCCCGUGCCAUGGCUUACCAGGCGUUGAGAAAUUCCGUAAA-GUGUUUUUACGGUAUUUGCGC------ ------------((((((.(((..............((((.........))))..(((.(((....))))))...))).))))))(((((-.(((....)))...)))))..------ ( -23.70, z-score = -0.08, R) >consensus ____________CGGGUCUGCCUGGUUUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAAAAGUGUUUUUACGGUAUUUGUGGGCUC__ ...................................(((((...(((.(((((.......(((....)))))))).)))..)))))......(((....)))................. (-14.36 = -14.21 + -0.15)
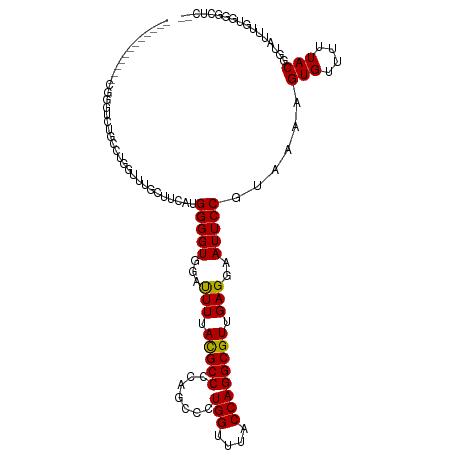

| Location | 4,221,247 – 4,221,354 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Shannon entropy | 0.53699 |
| G+C content | 0.49304 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4221247 107 - 21146708 --GAGCCAACAAAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAGACCAGGCAGCUCAGACC--------- --((((.........(((((.((....)).)))))...........(((((((....(((((.((((........)))).))).))......))))))).)))).....--------- ( -34.30, z-score = -3.42, R) >droSim1.chr2R 2882904 108 - 19596830 --GAGCCAACAAAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAGACCUGGCAGCCCAGACCC-------- --..((((.......(((((.((....)).)))))......(((...(((((....)))))..(((((.(.......).))))))))........))))...........-------- ( -28.60, z-score = -1.10, R) >droSec1.super_1 1867454 108 - 14215200 --GAGCCAACAAAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAGUCCACCCCAUGAAGCAGAACAGGCAGCCCAGACCC-------- --..(((........(((((.((....)).)))))......(((...(((((....)))))..(((((.(.......).)))))))).........)))...........-------- ( -28.00, z-score = -0.91, R) >droYak2.chr2L 16877015 98 - 22324452 --GAGCCAACAAAUACCGUAGAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAACCCAGGCC------------------ --..(((........(((((..........)))))......(((...(((((....)))))..(((((.(.......).)))))))).........))).------------------ ( -27.60, z-score = -1.65, R) >droEre2.scaffold_4929 8229677 116 + 26641161 --GAGCCAACAAAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAGCCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCGGACCAGGCAGCCCAGGCAGCCCAGACC --..(((........(((((.((....)).)))))...........(((((((..(((((((.((((........)))).))).))..))..)))))))......))).......... ( -37.90, z-score = -1.92, R) >droAna3.scaffold_13266 18111585 101 + 19884421 --GAGCCAACAAAUACCGUAGAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCAGGGGCGUUAAAUCCACCCCUUG--GCCAAGCAGCAAGUUCC------------- --((((.........(((((..........)))))...........((((((....))))((((((((.(.......).)))))..--)))..)).....)))).------------- ( -27.90, z-score = -1.34, R) >dp4.chr3 2202533 107 - 19779522 GUGCCAACAGAAAUACCGUAAAAACACUUUUUCGGAAUUCCUCGACGCCUGGUAGACCAGGGACGGGGCGUUAAAUCAACCCC-AGAGCCCAACAGAAAAAAAGGAAC---------- ...............(((.((((....)))).)))..(((((((.(.(((((....)))))).))((((..............-...))))...........))))).---------- ( -24.53, z-score = -0.37, R) >droPer1.super_2 2373915 107 - 9036312 GUGCCAACAGAAAUACCGUAAAAACACUUUUUCGGAAUUCCUCGACGCCUGGUAGACCAGGGACGGGGCGUUAAAUCAACCCC-AGAGCCCAACAGAAAAAAAGGAAC---------- ...............(((.((((....)))).)))..(((((((.(.(((((....)))))).))((((..............-...))))...........))))).---------- ( -24.53, z-score = -0.37, R) >droWil1.scaffold_180701 640680 87 - 3904529 ------GCAAAAAUACCGUAAAAACAC-UUUACGGAAUUUCUCAACGCCUGGUAAGCCAGGGACGGGGCGUAAAAUCAACCCC-AAAAAAAAGCU----------------------- ------((..((((.(((((((.....-))))))).)))).......(((((....)))))...((((...........))))-........)).----------------------- ( -24.40, z-score = -2.94, R) >droVir3.scaffold_12875 11359258 98 + 20611582 ------GCGCAAAUACCGUAAAAACAC-UUUACGGAAUUUCUCAACGCCUGGUAAGCCAUGGCACGGGCGUAAAAUCAACCCC-AAAACUUAACCAAAGUAACUCC------------ ------((((((((.(((((((.....-))))))).))))......((((((....))).)))....))))............-...((((.....))))......------------ ( -22.80, z-score = -2.33, R) >droMoj3.scaffold_6496 17883949 98 - 26866924 ------GCGCAAAUACCGUAAAAACAC-UUUACGGAAUUUCUCAACGCCUGGUAAGCCAUGGCACGGGCGUAAAAUCAACCCC-GAAACUUAACCAAAGUAACUCC------------ ------(.(.((((.(((((((.....-))))))).))))).)...((((((....))).))).((((.((.......)))))-)..((((.....))))......------------ ( -24.50, z-score = -2.57, R) >droGri2.scaffold_15245 15767445 99 + 18325388 ------GCGCAAAUACCGUAAAAACAC-UUUACGGAAUUUCUCAACGCCUGGUAAGCCAUGGCACGGGCAUAAAAUCAACCCCAAAAACUUAACCAAAGUAACUCC------------ ------((.(((((.(((((((.....-))))))).))))......((((((....))).)))..).))..................((((.....))))......------------ ( -21.40, z-score = -2.40, R) >consensus __GAGCCAACAAAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCAACCCCAUGAAGCAAACCAGACAAACCAG____________ ...............(((((..........))))).........((((((((....)).......))))))............................................... (-11.92 = -12.17 + 0.25)

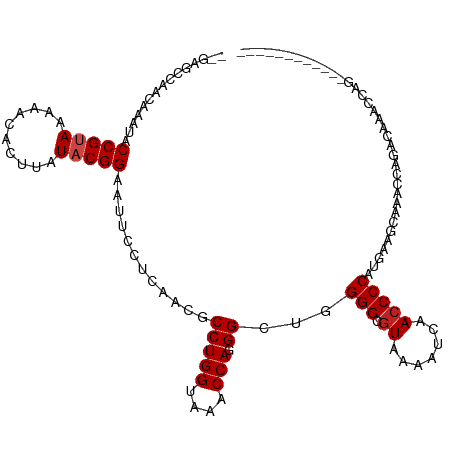

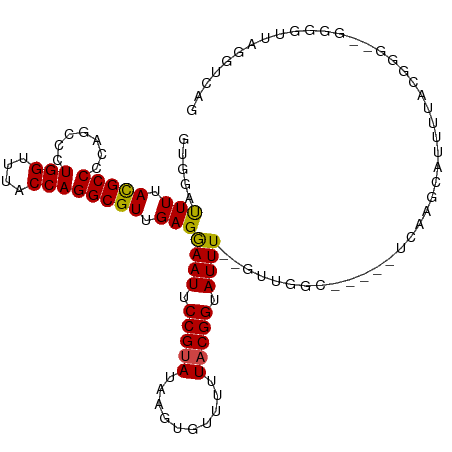
| Location | 4,221,276 – 4,221,381 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Shannon entropy | 0.43037 |
| G+C content | 0.48963 |
| Mean single sequence MFE | -36.59 |
| Consensus MFE | -13.41 |
| Energy contribution | -13.20 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4221276 105 + 21146708 GUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUU--GUUGGC-----UCAAGCAUUUUACGGG--GGGGUUAGGUCAG ...(((((.((.((((.(((.(((....))))))((..(.((((.(((((..........))))).))))--.)..))-----..............))--)).)).))))).. ( -37.60, z-score = -2.77, R) >droSim1.chr2R 2882934 104 + 19596830 GUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUU--GUUGGC-----UCAAGCAUUUUACGGG--GGG-UUAGGUCAG ...(((((.((.((((.(((.(((....))))))((..(.((((.(((((..........))))).))))--.)..))-----.............)))--).)-).))))).. ( -37.90, z-score = -3.12, R) >droSec1.super_1 1867484 103 + 14215200 GUGGACUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUU--GUUGGC-----UCAAGCAUUU-ACGGG--GGG-UUAGGUCAG ...(((((.((.((((.(((.(((....))))))((..(.((((.(((((..........))))).))))--.)..))-----..........-..)))--).)-).))))).. ( -40.30, z-score = -3.48, R) >droYak2.chr2L 16877035 104 + 22324452 GUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUCUACGGUAUUU--GUUGGC-----UCAAGCAUUUUACGGG--GGG-CUAGGUCAG ...(((((...(((((((((((((....))))).)))........(((((.((((((((....(((....--....))-----).))))))))))))))--)))-).))))).. ( -36.10, z-score = -1.97, R) >droEre2.scaffold_4929 8229715 104 - 26641161 GUGGAUUUUACGCCCCAGCCCUGGCUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUU--GUUGGC-----UCAAGCAUUUUACGGG--GGG-UUAGGUCAG ...(((((.((.((((.(((.(((....))))))((..(.((((.(((((..........))))).))))--.)..))-----.............)))--).)-).))))).. ( -37.10, z-score = -2.50, R) >droAna3.scaffold_13266 18111608 105 - 19884421 GUGGAUUUAACGCCCCUGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUCUACGGUAUUU--GUUGGC-----UCAAGCAUUUUCCGGG--GGGGUUAGGUCAG ...((((((((.((((((.(((((....))))).((..(.((((.(((((..........))))).))))--.)..))-----............))))--)).)))))))).. ( -42.50, z-score = -3.48, R) >dp4.chr3 2202560 109 + 19779522 GUUGAUUUAACGCCCCGUCCCUGGUCUACCAGGCGUCGAGGAAUUCCGAAAAAGUGUUUUUACGGUAUUUCUGUUGGC-----ACAAGUAUUUUUCCGGAUGGAGUUAGGUCAG ..(((((((((.((.((.((((((....))))).).)).))...((((((((((((((...((((.....)))).)))-----)).....))))).))))....))))))))). ( -35.50, z-score = -2.61, R) >droPer1.super_2 2373942 109 + 9036312 GUUGAUUUAACGCCCCGUCCCUGGUCUACCAGGCGUCGAGGAAUUCCGAAAAAGUGUUUUUACGGUAUUUCUGUUGGC-----ACAAGUAUUUUUCCGGAUGGAGUUAGGUCAG ..(((((((((.((.((.((((((....))))).).)).))...((((((((((((((...((((.....)))).)))-----)).....))))).))))....))))))))). ( -35.50, z-score = -2.61, R) >droWil1.scaffold_180701 640694 113 + 3904529 GUUGAUUUUACGCCCCGUCCCUGGCUUACCAGGCGUUGAGAAAUUCCGUA-AAGUGUUUUUACGGUAUUUUUGCCAGUGUUAUACGACGACGACUGGGUUAGGGGUUAGGUCAG ..((((((...(((((...(((((....))))).....((((((.(((((-(((...)))))))).)))))).((((((((.......))).)))))....))))).)))))). ( -37.50, z-score = -2.23, R) >droVir3.scaffold_12875 11359283 98 - 20611582 GUUGAUUUUACGCCCGUGCCAUGGCUUACCAGGCGUUGAGAAAUUCCGUA-AAGUGUUUUUACGGUAUUUGCGCCAGC-----UCCAGCUU----------GGGGUUAGGUCAG ..((((((.((.((((.((...((((.....(((((....((((.(((((-(((...)))))))).))))))))))))-----)...)).)----------))))).)))))). ( -32.30, z-score = -1.84, R) >droMoj3.scaffold_6496 17883974 97 + 26866924 GUUGAUUUUACGCCCGUGCCAUGGCUUACCAGGCGUUGAGAAAUUCCGUA-AAGUGUUUUUACGGUAUUUGCGCCAGC-----UCCAGCU-----------GGGGUUAGGUCAG ..((((((.((.((((.((...((((.....(((((....((((.(((((-(((...)))))))).))))))))))))-----)...)))-----------))))).)))))). ( -31.70, z-score = -1.63, R) >droGri2.scaffold_15245 15767471 98 - 18325388 GUUGAUUUUAUGCCCGUGCCAUGGCUUACCAGGCGUUGAGAAAUUCCGUA-AAGUGUUUUUACGGUAUUUGCGCCAGCUCC---UGGCUGGGGUUAGGUCAG------------ ..((((((...((((..((((.((((.....(((((....((((.(((((-(((...)))))))).)))))))))))).).---))))..)))).)))))).------------ ( -35.10, z-score = -2.40, R) >consensus GUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUUU__GUUGGC_____UCAAGCAUUUUACGGG__GGGGUUAGGUCAG .....(((.(((((.......(((....)))))))).)))((((.(((((..........))))).))))............................................ (-13.41 = -13.20 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:42 2011