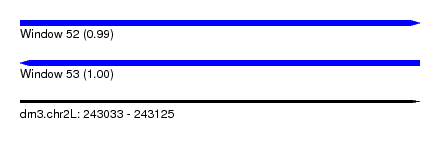
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 243,033 – 243,125 |
| Length | 92 |
| Max. P | 0.999712 |

| Location | 243,033 – 243,125 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Shannon entropy | 0.48007 |
| G+C content | 0.37428 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -13.94 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
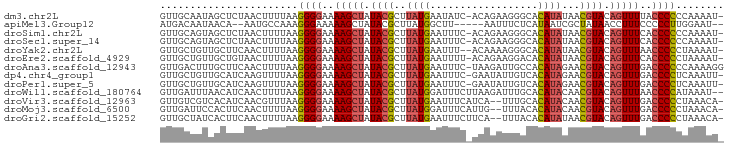
>dm3.chr2L 243033 92 + 23011544 GUUGCAAUAGCUCUAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUAUC-ACAGAAGGGCACAUAUAACGUACAGUUUUACCCCCCAAAAU- ...((....))............((((.((((((.((((..((((....((-......))...))))...)))).)))))).)))).......- ( -19.00, z-score = -1.39, R) >apiMel3.Group12 2847979 85 - 9182753 AUGACAAUAACA--AAUGCCAAAGGGAAAAAGCUAUACGCUUAUGGCUU-----AAUUUCUCAUAAUCGCUAUAACCUUUCCCCCUUGGAAU-- ............--....((((.((((((((((.....))))(((((..-----..............)))))....))))))..))))...-- ( -16.79, z-score = -1.78, R) >droSim1.chr2L 248258 92 + 22036055 GUUGCAGUAGCUCUAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUC-ACAGAAGGGCACAUAUAACGUACAGUUUCACCCCCCAAAAU- ...((....))............((((..(((((.((((..((((..((((-...))))....))))...)))).)))))..)))).......- ( -17.80, z-score = -0.61, R) >droSec1.super_14 241000 92 + 2068291 GUUGCAGUAGCUCUAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUC-ACAGAAGGGCACAUAUAACGUACAGUUUCACCCCCCAAAAU- ...((....))............((((..(((((.((((..((((..((((-...))))....))))...)))).)))))..)))).......- ( -17.80, z-score = -0.61, R) >droYak2.chr2L 236053 91 + 22324452 GUUGCUGUUGCUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUU--ACAAAAGGGCACAUAUAACGUACAGUUUAACCCCCUAAAAU- ...((....))......(((((.((((..(((((.((((..((((..(((--......)))..))))...)))).)))))..)))).))))).- ( -18.60, z-score = -0.91, R) >droEre2.scaffold_4929 289350 92 + 26641161 GUUGCUGUUGCUGUAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUU-ACAGAAGGACACAUAUAACGUACAGUUUCACCCCCUAAAAU- (((((.......)))))(((((.((((..(((((.((((..((((..((((-.....))))..))))...)))).)))))..)))).))))).- ( -23.10, z-score = -2.46, R) >droAna3.scaffold_12943 891856 93 - 5039921 GUUGACUUUGCUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUC-UAAGAUUGCCACAUAGAACGUACAGUUUGACCCCCCAAAAGG ................(((((..((((..(((((.((((.(((((((((..-...))))....)))))..)))).)))))..))))..))))). ( -20.00, z-score = -1.44, R) >dp4.chr4_group1 916627 92 - 5278887 GUUGCUGUUGCAUCAAGUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUC-GAAUAUUGUCACAUAGAACGUACAGUUUGACCCCUCAAAUU- (.(((....))).)....(((.(((((..(((((.((((.(((((......-...........)))))..)))).)))))..))))).)))..- ( -20.23, z-score = -1.35, R) >droPer1.super_5 848179 92 + 6813705 GUUGCUGUUGCAUCAAGUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUC-GAAUAUUGUCACAUAGAACGUACAGUUUGACCCCUCAAAUU- (.(((....))).)....(((.(((((..(((((.((((.(((((......-...........)))))..)))).)))))..))))).)))..- ( -20.23, z-score = -1.35, R) >droWil1.scaffold_180764 130107 92 + 3949147 GUUGAUUUAACAUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGGAUUUCUUAAGAUUUGCACAUACAACGUACAGUUUAACCCCAUAAAU-- ((((((.....)))))).((((.((((..(((((.((((..(((((...((....))....).))))...)))).)))))..)))).)))).-- ( -22.70, z-score = -4.12, R) >droVir3.scaffold_12963 11349742 91 - 20206255 GUUGUCGUCACAUCAACGUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCAUCA--UUUGCACAUACAACGUACAGUUUGACCCCCUAAACA- ((((.........))))(((((.((((..(((((.(((((..((((......)))--)..)).........))).)))))..)))).))))).- ( -23.70, z-score = -3.48, R) >droMoj3.scaffold_6500 2474493 91 + 32352404 GUUGAUUCCACUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGGAUUUCAUUG--UUUACACAUACAACGUACAGUUUGACCCCCUAAACA- (((((.......))))).((((.((((..(((((.((((..((((..........--......))))...)))).)))))..)))).))))..- ( -22.19, z-score = -3.35, R) >droGri2.scaffold_15252 12934001 91 + 17193109 GUUGCUAUCACUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCUUCA--UUUACACAUAUAACGUACAGUUUGACCCCCUAAACA- ((((.........)))).((((.((((..(((((.((((..((((..........--......))))...)))).)))))..)))).))))..- ( -18.09, z-score = -2.97, R) >consensus GUUGCUGUAGCUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUC_ACAGAUUGGCACAUAUAACGUACAGUUUGACCCCCCAAAAU_ .......................((((..(((((.((((..((((..................))))...)))).)))))..))))........ (-13.94 = -14.19 + 0.25)
| Location | 243,033 – 243,125 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Shannon entropy | 0.48007 |
| G+C content | 0.37428 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.29 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
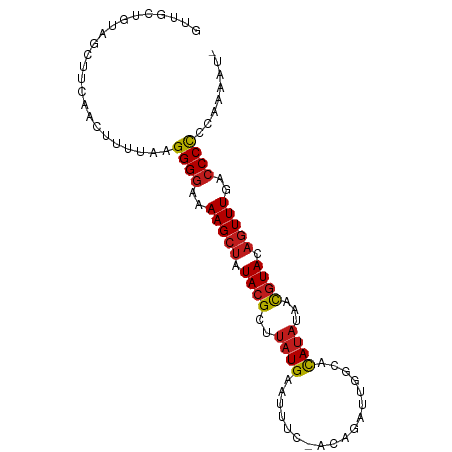
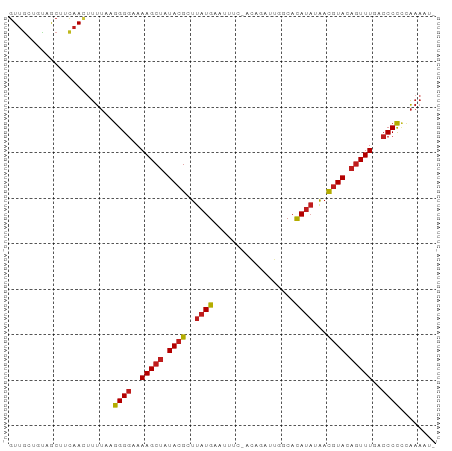
>dm3.chr2L 243033 92 - 23011544 -AUUUUGGGGGGUAAAACUGUACGUUAUAUGUGCCCUUCUGU-GAUAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUAGAGCUAUUGCAAC -.((((((((((.(((((((((((((.((((...........-......))))))))))))).))))))))))))))......((....))... ( -27.73, z-score = -3.06, R) >apiMel3.Group12 2847979 85 + 9182753 --AUUCCAAGGGGGAAAGGUUAUAGCGAUUAUGAGAAAUU-----AAGCCAUAAGCGUAUAGCUUUUUCCCUUUGGCAUU--UGUUAUUGUCAU --...(((((((((((((((((((.((.(((((.(.....-----...)))))).)))))))))))))))))))))....--............ ( -30.50, z-score = -4.68, R) >droSim1.chr2L 248258 92 - 22036055 -AUUUUGGGGGGUGAAACUGUACGUUAUAUGUGCCCUUCUGU-GAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUAGAGCUACUGCAAC -.((((((((((.(((.(((((((((.((((...........-......))))))))))))).))).))))))))))......((....))... ( -28.03, z-score = -2.98, R) >droSec1.super_14 241000 92 - 2068291 -AUUUUGGGGGGUGAAACUGUACGUUAUAUGUGCCCUUCUGU-GAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUAGAGCUACUGCAAC -.((((((((((.(((.(((((((((.((((...........-......))))))))))))).))).))))))))))......((....))... ( -28.03, z-score = -2.98, R) >droYak2.chr2L 236053 91 - 22324452 -AUUUUAGGGGGUUAAACUGUACGUUAUAUGUGCCCUUUUGU--AAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGCAACAGCAAC -.((((((((((..((.(((((((((.(((((((......))--)....))))))))))))).))..))))))))))((((......))))... ( -29.40, z-score = -4.23, R) >droEre2.scaffold_4929 289350 92 - 26641161 -AUUUUAGGGGGUGAAACUGUACGUUAUAUGUGUCCUUCUGU-AAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUACAGCAACAGCAAC -.((((((((((.(((.(((((((((.((((...........-......))))))))))))).))).))))))))))......((....))... ( -27.93, z-score = -3.98, R) >droAna3.scaffold_12943 891856 93 + 5039921 CCUUUUGGGGGGUCAAACUGUACGUUCUAUGUGGCAAUCUUA-GAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGCAAAGUCAAC .(((((((((((..((.(((((((((.((((...........-......))))))))))))).))..)))))))))))................ ( -27.43, z-score = -3.25, R) >dp4.chr4_group1 916627 92 + 5278887 -AAUUUGAGGGGUCAAACUGUACGUUCUAUGUGACAAUAUUC-GAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAACUUGAUGCAACAGCAAC -..(((((((((..((.(((((((((...(((((........-.....)))))))))))))).))..)))))))))......(((....))).. ( -27.02, z-score = -4.21, R) >droPer1.super_5 848179 92 - 6813705 -AAUUUGAGGGGUCAAACUGUACGUUCUAUGUGACAAUAUUC-GAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAACUUGAUGCAACAGCAAC -..(((((((((..((.(((((((((...(((((........-.....)))))))))))))).))..)))))))))......(((....))).. ( -27.02, z-score = -4.21, R) >droWil1.scaffold_180764 130107 92 - 3949147 --AUUUAUGGGGUUAAACUGUACGUUGUAUGUGCAAAUCUUAAGAAAUCCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAUGUUAAAUCAAC --.((((.((((..((.(((((((((.((((.(....((....))...)))))))))))))).))..)))).)))).((((((.....)))))) ( -26.40, z-score = -4.62, R) >droVir3.scaffold_12963 11349742 91 + 20206255 -UGUUUAGGGGGUCAAACUGUACGUUGUAUGUGCAAA--UGAUGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAACGUUGAUGUGACGACAAC -.((((((((((..((.(((((((((((....))..(--(((......)))).))))))))).))..))))))))))((((......))))... ( -31.20, z-score = -4.35, R) >droMoj3.scaffold_6500 2474493 91 - 32352404 -UGUUUAGGGGGUCAAACUGUACGUUGUAUGUGUAAA--CAAUGAAAUCCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGUGGAAUCAAC -..(((((((((..((.(((((((((...(((....)--))(((.....))).))))))))).))..))))))))).(((((.......))))) ( -26.80, z-score = -3.63, R) >droGri2.scaffold_15252 12934001 91 - 17193109 -UGUUUAGGGGGUCAAACUGUACGUUAUAUGUGUAAA--UGAAGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGUGAUAGCAAC -..(((((((((..((.(((((((((.((....)).(--((((....))))).))))))))).))..))))))))).((((......))))... ( -26.10, z-score = -3.90, R) >consensus _AUUUUAGGGGGUCAAACUGUACGUUAUAUGUGCCAAUCUGA_GAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGCAACAGCAAC ...(((((((((..((.(((((((((.((((..................))))))))))))).))..))))))))).................. (-21.94 = -22.29 + 0.35)


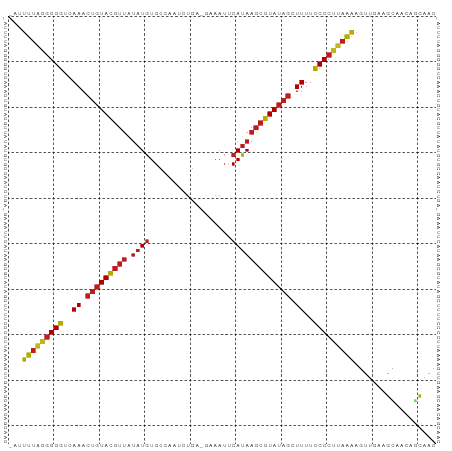
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:23 2011