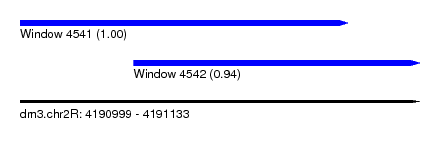
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,190,999 – 4,191,133 |
| Length | 134 |
| Max. P | 0.999213 |

| Location | 4,190,999 – 4,191,109 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.51132 |
| G+C content | 0.36412 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.41 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
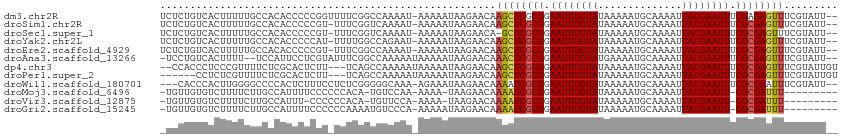
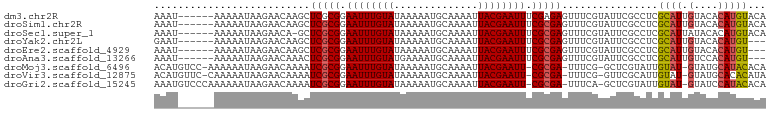
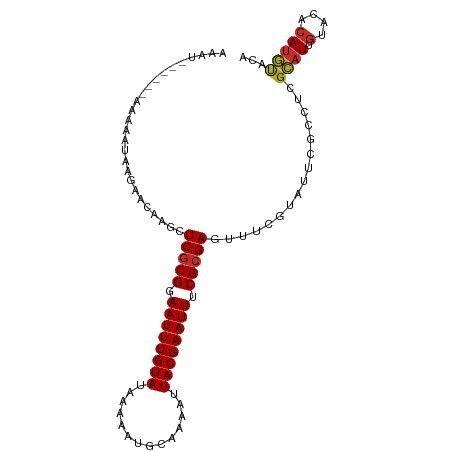
>dm3.chr2R 4190999 110 + 21146708 UCUCUGUCACUUUUUGCCACACCCCCGGUUUUCGGCCAAAAU-AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGAGAGUUUCGUAUU-- ..(((......(((((((..(((...)))....)).))))).-.......)))((((((((.(((((((((((..............))))))))))).)))))).))...-- ( -23.38, z-score = -1.68, R) >droSim1.chr2R 2850882 109 + 19596830 UCUCUGUCACUUUUUGCCACACCCCCGU-UUUCGGUCAAAAU-AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUU-- ..(((......(((((((..........-....)).))))).-.......)))((((((((((((((((((((..............)))))))))))))))))).))...-- ( -27.62, z-score = -3.64, R) >droSec1.super_1 1837540 108 + 14215200 UCUCUGUCACUUUUUGCCACACCCCCGU-UUUCGGUCAAAAU-AAAAAUAAGAACA-GCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUU-- ..(((......(((((((..........-....)).))))).-.......)))..(-((((((((((((((((..............))))))))))))))))).......-- ( -25.62, z-score = -2.96, R) >droYak2.chr2L 16846033 109 + 22324452 UCUCUGUCACUUUUUGCCACACCCCCAU-UUUUGGCCAGAAU-AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUU-- ..(((......((((((((.........-...))).))))).-.......)))((((((((((((((((((((..............)))))))))))))))))).))...-- ( -28.68, z-score = -3.93, R) >droEre2.scaffold_4929 8199567 109 - 26641161 UCUCUGUCACUUUUUGCCACACCCCCGU-UUUCGGCCAAAAU-AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUU-- ..(((......(((((((..........-....)).))))).-.......)))((((((((((((((((((((..............)))))))))))))))))).))...-- ( -27.62, z-score = -3.53, R) >droAna3.scaffold_13266 18082804 108 - 19884421 -UCCUGUCACUUUU--UCCAUUCCUCGUAUUUCGGCCAAAAAUAAAAAUAAGAACAAACUCGCGGAAUUUGUAUGAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUU-- -.............--.....................................((((((((((((((((((((..............)))))))))))))))))).))...-- ( -23.84, z-score = -2.55, R) >dp4.chr3 2175155 108 + 19779522 --CCACCCUCCCGUUUUCUCGCACUCUU---UCAGCCAAAAAUAAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUGU --..........(((((...((......---...))..)))))....((((..((((((((((((((((((((..............)))))))))))))))))).)).)))) ( -24.04, z-score = -3.21, R) >droPer1.super_2 2345041 104 + 9036312 ------CCUCUCGUUUUCUCGCACUCUU---UCAGCCAAAAAUAAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUGU ------..(((..((((...........---.............))))..)))((((((((((((((((((((..............)))))))))))))))))).))..... ( -24.30, z-score = -2.90, R) >droWil1.scaffold_180701 598341 107 + 3904529 ---CACCCACUUGGGGCCCCACUCUUUCCUCUCGGGGGCAAA-AGAAAUAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAAUUUCGUAUU-- ---..(((....)))(((((..............)))))...-.(((((..........((((((((((((((..............))))))))))))))))))).....-- ( -32.18, z-score = -3.49, R) >droMoj3.scaffold_6496 8939435 99 - 26866924 -UGUUGUGUCUUUUCUUGCCAUUUUCCCCCCACA-UGUCCAA-AAAA-UAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUU-CGCGAUUU--------- -..((((.((((...(((.(((...........)-))..)))-....-.))))))))(((((((.((((((((..............))))))))-))))))).--------- ( -16.44, z-score = -1.24, R) >droVir3.scaffold_12875 9212695 98 + 20611582 -UGUUGUGUCUUUUCUUGCCAUUU-CCCCCCACA-UGUUCCA-AAAA-UAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUU-CGCGAUUU--------- -...((((................-.....))))-(((((..-....-...)))))((((((((.((((((((..............))))))))-))))))))--------- ( -18.44, z-score = -2.03, R) >droGri2.scaffold_15245 2524647 101 + 18325388 -UGUUGUGUCUUUUCUUGCCAUUUUCCCCCCAAAAUGUCCCA-AAAAAUAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUU-CGCGAUUU--------- -..((((.((((...(((.((((((......))))))...))-).....))))))))(((((((.((((((((..............))))))))-))))))).--------- ( -19.54, z-score = -2.69, R) >consensus _CUCUGUCACUUUUUGCCACACCCUCGU_UUUCGGCCAAAAA_AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUU__ ........................................................((((((((.((((((((..............)))))))).))))))))......... (-16.74 = -16.41 + -0.33)
| Location | 4,191,037 – 4,191,133 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.26 |
| Shannon entropy | 0.27891 |
| G+C content | 0.33820 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941300 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 4191037 96 + 21146708 AAAU------AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGAGAGUUUCGUAUUCGCCUCGCAUUGUACACAUGUACA ....------..........((((((((.(((((((((((..............))))))))))).)))))).)).............(((((....))))) ( -21.84, z-score = -2.32, R) >droSim1.chr2R 2850919 96 + 19596830 AAAU------AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUCGCCUCGCAUUGUACACAUGUACA ....------..........((((((((((((((((((((..............)))))))))))))))))).)).............(((((....))))) ( -27.54, z-score = -4.00, R) >droSec1.super_1 1837577 95 + 14215200 AAAU------AAAAAUAAGAACA-GCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUCGCCUCGCAUUAUACACAUGUACA ....------........(((..-((((((((((((((((..............)))))))))))))))))))((((..((...))...))))......... ( -24.74, z-score = -3.42, R) >droYak2.chr2L 16846070 93 + 22324452 GAAU------AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUCGCCUCGCAUUGUACACAUGU--- ....------............((((((((((((((((((..............)))))))))))))))))).((((..((...))...))))......--- ( -25.04, z-score = -3.09, R) >droEre2.scaffold_4929 8199604 93 - 26641161 AAAU------AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUCGCCUCGCAUUGUACACAUGU--- ....------............((((((((((((((((((..............)))))))))))))))))).((((..((...))...))))......--- ( -25.04, z-score = -3.28, R) >droAna3.scaffold_13266 18082840 93 - 19884421 AAAU------AAAAAUAAGAACAAACUCGCGGAAUUUGUAUGAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUCGCCUCGCAUUGUCCACAUGU--- ....------..........((((((((((((((((((((..............)))))))))))))))))).)).........((((.(....)))))--- ( -23.94, z-score = -2.87, R) >droMoj3.scaffold_6496 8939465 97 - 26866924 ACAUGUCC-AAAAAAUAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUU-CGCGA-UUUCG-GCUCGUAUUGUAU-GUAUGCAUACACA ........-....((((.((.(.((((((((.((((((((..............))))))))-)))))-))).)-..)).))))((((-(....)))))... ( -23.04, z-score = -2.41, R) >droVir3.scaffold_12875 9212724 97 + 20611582 ACAUGUUC-CAAAAAUAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUU-CGCGA-UUUCG-GUUCGCAUUGUAU-GUAUGCACACAUA ..((((..-.........((((.((((((((.((((((((..............))))))))-)))))-)))..-))))((((.....-..))))..)))). ( -25.14, z-score = -2.90, R) >droGri2.scaffold_15245 2524678 98 + 18325388 AAAUGUCCCAAAAAAUAAGAACAAAAUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUU-CGCGA-UUUCA-GCUCGUAUUGUAU-GUAUCCAUACACA .............((((.((.(.((((((((.((((((((..............))))))))-)))))-)))..-).)).))))((((-(....)))))... ( -20.34, z-score = -2.74, R) >consensus AAAU______AAAAAUAAGAACAAGCUCGCGGAAUUUGUAUAAAAAUGCAAAAUUACGAAUUUCGCGAGUUUCGUAUUCGCCUCGCAUUGUACACAUGUACA ..................(((.....(((((.((((((((..............)))))))).)))))..)))...........((((.......))))... (-12.87 = -12.43 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:38 2011