| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,188,971 – 4,189,066 |
| Length | 95 |
| Max. P | 0.998596 |

| Location | 4,188,971 – 4,189,066 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.42 |
| Shannon entropy | 0.67641 |
| G+C content | 0.46698 |
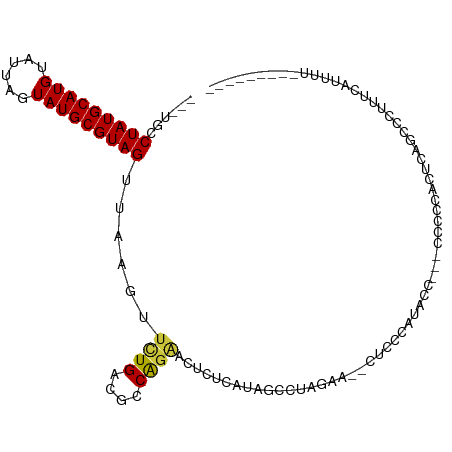
| Mean single sequence MFE | -15.93 |
| Consensus MFE | -10.71 |
| Energy contribution | -10.67 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
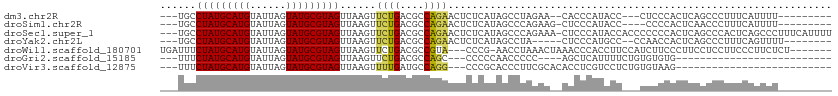
>dm3.chr2R 4188971 95 + 21146708 ---UGCCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCAGAACUCUCAUAGCCUAGAA--CACCCAUACC---CUCCCACUCAGCCCUUUCAUUUU--------- ---.(((((((((((......)))))))))...(((((((....)))))))......))......--..........---.......................--------- ( -17.90, z-score = -2.22, R) >droSim1.chr2R 2848914 95 + 19596830 ---UGCCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCAGAACUCUCAUAGCCCAGAAG-CUCCCAUACC----CCCCACUCAACCCUUUCAUUUU--------- ---.(((((((((((......)))))))))...(((((((....)))))))..............)-).........----......................--------- ( -18.30, z-score = -2.79, R) >droSec1.super_1 1835591 108 + 14215200 ---UGCCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCAGAACUCUCAUAGCCCAGAAA-CUCCCAUACCACCCCCCCACUCAGCCCACUCAGCCCUUUCAUUUU ---.(((((((((((......)))))))))...(((((((....)))))))......)).......-............................................. ( -17.90, z-score = -2.20, R) >droYak2.chr2L 16844110 94 + 22324452 ---UGCCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCAGAACUCUCAUAGCCUA-----CUCCCAUGCC--CCAACCACUCAGCCCUUUCAGUUUU-------- ---.......(((((....((((.((.......(((((((....)))))))......)).))-----))..))))).--.........................-------- ( -18.52, z-score = -1.96, R) >droWil1.scaffold_180701 594995 101 + 3904529 UGAUUUCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCGUA---CCCG-AACCUAAACUAAACCCACCUUCCAUCUUCCCUUCCUCCUUCCCUUCUCU------- ......(((((((((......)))))))))(((.((((.(........---.).)-))).)))..........................................------- ( -11.30, z-score = -1.57, R) >droGri2.scaffold_15185 520 76 + 3138 ---UUUCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCAGC---CCCCCAACCCCC----AGCUCAUUUUCUGUGUGUG-------------------------- ---...(((((((((......)))))))))..........(((((((.---............----..........))).)))).-------------------------- ( -13.00, z-score = -1.51, R) >droVir3.scaffold_12875 9209398 80 + 20611582 ---UUUCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUUUGAUGCCAGG---CCCGCACCCUUCGCACACCUCGUCCUCUGUGUAAG-------------------------- ---...(((((((((......))))))))).........((((..(((---...((.......))...)))))))...........-------------------------- ( -14.60, z-score = -0.14, R) >consensus ___UGCCUAUGCAUGUAUUAGUAUGCGUAGUUAAGUUCUGACGCCAGAACUCUCAUAGCCUAGAA__CUCCCAUACC___CCCCCACUCAGCCCUUUCAUUUU_________ ......(((((((((......)))))))))......((((....))))................................................................ (-10.71 = -10.67 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:36 2011