| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,178,313 – 4,178,413 |
| Length | 100 |
| Max. P | 0.836415 |

| Location | 4,178,313 – 4,178,413 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Shannon entropy | 0.32291 |
| G+C content | 0.51111 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -19.65 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4178313 100 + 21146708 ----------------GCCAUGACUCCGGAAACGGAAGUGCCGCUGUCAC-AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAA-GUUGCCCUUUGCCC-- ----------------(.(((...((((....)))).))).)((.(((.(-(..((((.((((((.(......).))))))))))...)).))).)).(((((-(.....))))))..-- ( -30.00, z-score = -1.76, R) >droSim1.chr2R 2836596 100 + 19596830 ----------------GCCAUGACUCCGGAAACGGAAGUGCCGCUGUCAC-AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAA-GUUGCCCUUUGCCC-- ----------------(.(((...((((....)))).))).)((.(((.(-(..((((.((((((.(......).))))))))))...)).))).)).(((((-(.....))))))..-- ( -30.00, z-score = -1.76, R) >droSec1.super_1 1825045 98 + 14215200 ----------------GCCAUGACUGCGGAAACGGAAGUGCCGCUGUC---AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAA-GUUGCCCUUUGCCC-- ----------------...(((((.((((..((....)).)))).)))---)).((((.((((((.(......).)))))))))).............(((((-(.....))))))..-- ( -32.50, z-score = -2.36, R) >droYak2.chr2L 16833254 100 + 22324452 ----------------GCCAUGACUCCGGAAACGGAAGUGCCGCUGUCAC-AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAA-GUUGCCCUUUUCCC-- ----------------((......((((....)))).(((((((.((((.-....))))))))).))....)).........((((((((.........))))-))))..........-- ( -29.10, z-score = -1.76, R) >droEre2.scaffold_4929 8187348 100 - 26641161 ----------------GCCAUGACUCCGGAAACGGAAGUGCCGCUGUCAC-AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAA-GUUGCCCUUUGCCC-- ----------------(.(((...((((....)))).))).)((.(((.(-(..((((.((((((.(......).))))))))))...)).))).)).(((((-(.....))))))..-- ( -30.00, z-score = -1.76, R) >droAna3.scaffold_13266 18070321 103 - 19884421 ----------------GGCCCGACUCCGGAAACGGAAGUGCCGCUGUCAC-AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAAAGUUGCCCUUUGGACCC ----------------((.((((.((((....))))..(((..(.(((.(-(..((((.((((((.(......).))))))))))...)).))).)..)))...........)))).)). ( -33.00, z-score = -2.03, R) >dp4.chr3 2163055 98 + 19779522 ----------------GCCAUGACACCGGAAACGGAAGUGCCGCUGUCAG-AUAUUGACAUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCACA-GCUUCCCGUCUC---- ----------------.....(((.(((....)))..((((..(.(((.(-(..((((.((((((.(......).))))))))))..))..))).)..)))).-.......)))..---- ( -32.70, z-score = -2.80, R) >droPer1.super_2 2332673 98 + 9036312 ----------------GCCAUGACACCGGAAACGGAAGUGCCGCUGUCAG-AUAUUGACAUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCACA-GCUUCCCUUCUC---- ----------------((.......(((....)))..((((..(.(((.(-(..((((.((((((.(......).))))))))))..))..))).)..)))).-))..........---- ( -30.50, z-score = -2.53, R) >droVir3.scaffold_12875 9196356 111 + 20611582 AGCAGCUAUGCAGUUAGCCAUGACACCGGAAACGGAAGUGCUGCUGUCACCAUAUUGACAUGGCGACAAAUGCG-CUCCGCUCAAGUU-GAGAUUGUAGCAAA---UGGCAUUCAA---- .(((....))).....(((((....(((....)))...((((((((((.((((......)))).))))......-(((.((....)).-)))...)))))).)---))))......---- ( -38.10, z-score = -2.04, R) >droMoj3.scaffold_6496 8918292 104 - 26866924 -------AGUCAGUACGCCAUGACACCGGAAACGGAAGUGCUGCUGUCACCAUAUUGACAUGGCGACAAAUGCG-CUCCGCUCAAGUU-GAGAUUGUAGCAAA---UGGCUUUCAA---- -------.........(((((....(((....)))...((((((((((.((((......)))).))))......-(((.((....)).-)))...)))))).)---))))......---- ( -34.80, z-score = -2.05, R) >droGri2.scaffold_15245 2511988 104 + 18325388 -------AGCAACGAUGCCAUGACACCGGAAACGGAAGUGCUGCUGUCACCAUAUUGACAUGGCGACAAAUGCG-CUCCGCUCAAGUA-GAGAUUGUAGCAAA---UGGCUUUCAA---- -------......((.(((((....(((....)))...((((((((((.((((......)))).))))......-(((..(....)..-)))...)))))).)---))))..))..---- ( -36.20, z-score = -2.76, R) >consensus ________________GCCAUGACUCCGGAAACGGAAGUGCCGCUGUCAC_AUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAA_GUUGCCCUUUGC_C__ ................(((((....(((....)))..........((((......)))))))))......(((.......(((......)))......)))................... (-19.65 = -19.48 + -0.16)

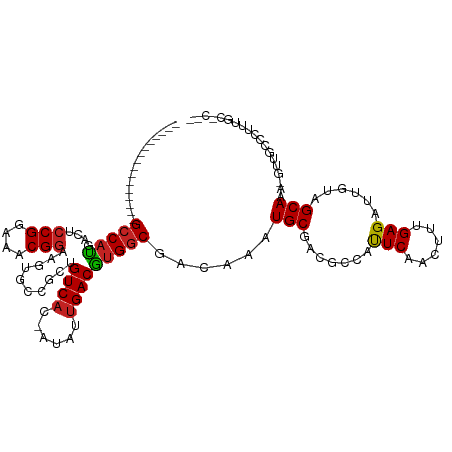

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:34 2011