
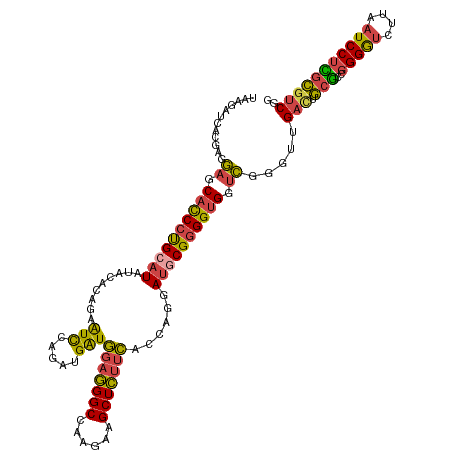
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,147,014 – 4,147,170 |
| Length | 156 |
| Max. P | 0.747443 |

| Location | 4,147,014 – 4,147,134 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Shannon entropy | 0.39272 |
| G+C content | 0.56528 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -31.28 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
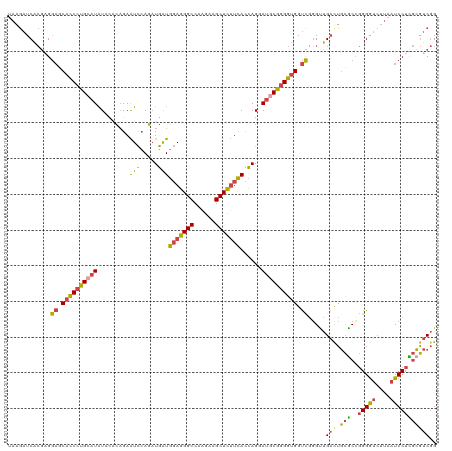
>dm3.chr2R 4147014 120 - 21146708 UAAGAUCACGAGGAGCACCCUGCAUGUACACAGAAUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGUGGUCGGGUUGAUUGCGCCGGGGUCUUAAUCCUCGCGUCGG ...((.(.((((((.((((((((.((....))..((((...((.((((((((......)))))))).)))))))))))))).((((((.....)).))))........)))))).))).. ( -46.20, z-score = -1.50, R) >droSec1.super_1 1793264 120 - 14215200 UAAGAUCACGAGGAGCACCCUGCAUGUACACAGAAUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGUGGUCGGGUUGACUGCGCCGGGGUCUUAAUCCUCGCGUCGG ...((.(.((((((.((((((((.((....))..((((...((.((((((((......)))))))).)))))))))))))).((((((.....)).))))........)))))).))).. ( -46.20, z-score = -1.30, R) >droSim1.chr2R 2813328 120 - 19596830 UAAGAUCACGGGGAGCACCCUGCAUGUACACAGAAUCCAGAUGGUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGUGGUCGGGUUGACUGCGCCGGGGUCUUAAUCCUCGCGUCGG ...((.(.((((((.((((((((.((....))..((((...(((((((((((......))))))))))))))))))))))).((((((.....)).))))........)))))).))).. ( -51.90, z-score = -2.36, R) >droYak2.chr2L 16798658 120 - 22324452 UAAGAUCGCGAGGAGCACCCUGCAUAUAAACAGAAUCCAGAUGAUGGAGGGCCAGGAAGCUCUUCACCAGUAUGCGGGGUGGUCGGGAUGACUGCUCUGGGAUCUUCAUCCUGGCGUCGG ...((.(((...((.((((((((((((.....(....)...((.((((((((......)))))))).)))))))))))))).))(((((((...(.....)....))))))).))))).. ( -47.10, z-score = -1.85, R) >droEre2.scaffold_4929 8155379 120 + 26641161 UAAGAUCACGAGGAGCACCCUGCAUAUAAACAGAGUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGAAUACGGGGUGGUCGGGCUGACUGCGCUGGGGUCUUAAUCCUUGCGUCGG ...((.(.((((((((.....))........(((.(((((.((.((((((((......)))))))).)).........(..(((.....)))..).))))).)))...)))))).))).. ( -39.00, z-score = -0.21, R) >droAna3.scaffold_13266 18041571 120 + 19884421 UAAACUCGAUCGACACCUCUCGCAUAUACGACGAGUUAAUUUGAUUCAAGGCCUUGAAGCUUCUGAUUAGCAGCCGUGGCGGCUUCGUUGUCUCUGCCCGGGAGAAGGUCCUCCGAUCCG .......(((((..((((((((((.(.((((((((((((((.(((((((....)))))...)).)))))))(((((...)))))))))))).).)))....))).))))....))))).. ( -36.70, z-score = -0.88, R) >consensus UAAGAUCACGAGGAGCACCCUGCAUAUACACAGAAUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGUGGUCGGGUUGACUGCGCCGGGGUCUUAAUCCUCGCGUCGG ............((.((((((((((.........(((.....)))(((((((......)))))))......)))))))))).)).....(((.(((..(((((....))))))))))).. (-31.28 = -30.93 + -0.35)

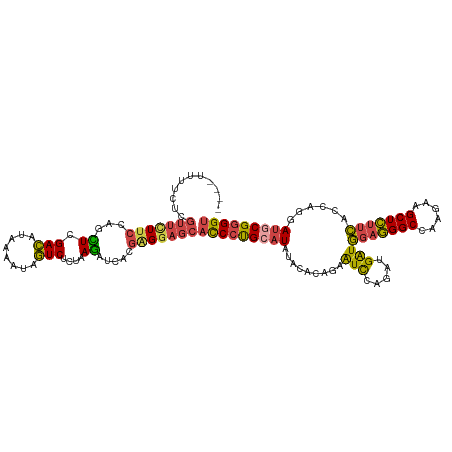
| Location | 4,147,054 – 4,147,170 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.40224 |
| G+C content | 0.47318 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
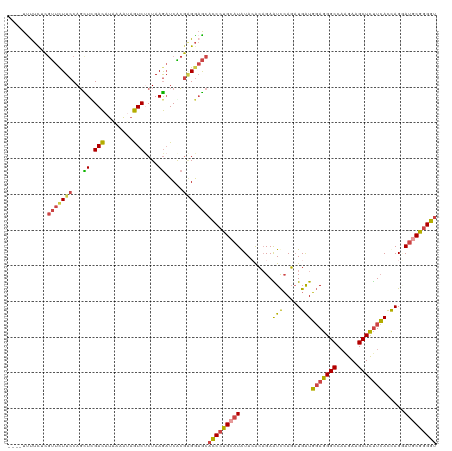
>dm3.chr2R 4147054 116 - 21146708 ----UUCUCUCGUUUUUCCAGCUUGACAUAAAAUAGUCUCUAAGAUCACGAGGAGCACCCUGCAUGUACACAGAAUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGU ----(((.(((((..((..((...(((........))).))..))..)))))))).(((((((.((....))..((((...((.((((((((......)))))))).))))))))))))) ( -35.30, z-score = -1.23, R) >droSec1.super_1 1793304 116 - 14215200 ----UUUUCUCGUUCUUCCAGCUCGACAUAAAAUAGUCUCUAAGAUCACGAGGAGCACCCUGCAUGUACACAGAAUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGU ----.((((((((((((..((...(((........))).))))))..)))))))).(((((((.((....))..((((...((.((((((((......)))))))).))))))))))))) ( -36.20, z-score = -1.52, R) >droSim1.chr2R 2813368 116 - 19596830 ----UUUUCUCGUUCUUCCAGCUCGACAUAAAAUAGUCUCUAAGAUCACGGGGAGCACCCUGCAUGUACACAGAAUCCAGAUGGUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGU ----.((((((((((((..((...(((........))).))))))..)))))))).(((((((.((....))..((((...(((((((((((......)))))))))))))))))))))) ( -41.90, z-score = -2.59, R) >droYak2.chr2L 16798698 116 - 22324452 ----UUUUUGCGUUUUUCCAGCUUGACAUAAAAAAGUCUCUAAGAUCGCGAGGAGCACCCUGCAUAUAAACAGAAUCCAGAUGAUGGAGGGCCAGGAAGCUCUUCACCAGUAUGCGGGGU ----((((((((.((((..(((((.........))).))..)))).))))))))..(((((((((((.....(....)...((.((((((((......)))))))).))))))))))))) ( -36.50, z-score = -1.96, R) >droEre2.scaffold_4929 8155419 116 + 26641161 ----UCUUCGCGUUCUUCCAGUUUGACAUAAAAAAGUCCCUAAGAUCACGAGGAGCACCCUGCAUAUAAACAGAGUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGAAUACGGGGU ----((((((.(.((((..((...(((........))).)))))).).))))))..((((((..........(....)...((.((((((((......)))))))).)).....)))))) ( -28.50, z-score = -0.26, R) >droAna3.scaffold_13266 18041611 120 + 19884421 UGAAUUUACAACCACUUCCAUUUCGAUAAGAAUUGAUCACUAAACUCGAUCGACACCUCUCGCAUAUACGACGAGUUAAUUUGAUUCAAGGCCUUGAAGCUUCUGAUUAGCAGCCGUGGC ...........((((......(((((...(..(((((((...((((((.(((................)))))))))....))).))))..).)))))(((.......)))....)))). ( -21.69, z-score = -0.15, R) >consensus ____UUUUCUCGUUCUUCCAGCUCGACAUAAAAUAGUCUCUAAGAUCACGAGGAGCACCCUGCAUAUACACAGAAUCCAGAUGAUGGAGGGCCAAGAAGCUCUUCACCAGGAUGCGGGGU ...........(((((((...((.(((........)))....)).....)))))))(((((((((.........(((.....)))(((((((......)))))))......))))))))) (-22.77 = -22.70 + -0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:32 2011