| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,140,777 – 4,140,871 |
| Length | 94 |
| Max. P | 0.815383 |

| Location | 4,140,777 – 4,140,871 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.37 |
| Shannon entropy | 0.55067 |
| G+C content | 0.47121 |
| Mean single sequence MFE | -16.91 |
| Consensus MFE | -9.99 |
| Energy contribution | -10.26 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
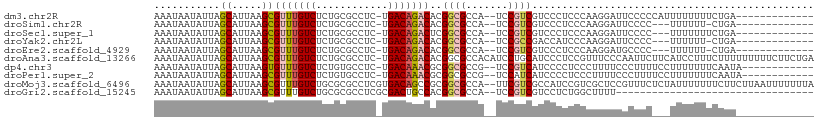
>dm3.chr2R 4140777 94 + 21146708 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC-UGACAGACACGGCGCCA--UCCGUCGUCCCUCCCAAGGAUUCCCCCAUUUUUUUUCUGA------------- ...........((.....))(((((((..........-.)))))))((((((...--..)))))).(((....))).....................------------- ( -16.80, z-score = -0.67, R) >droSim1.chr2R 2807520 90 + 19596830 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC-UGACAGACACGGCGCCA--UCCGUCGUCCCUCCCAAGGAUUCCCC---UUUUUU-CUGA------------- ...........((.....))(((((((..........-.)))))))((((((...--..)))))).......((((.....))---))....-....------------- ( -17.50, z-score = -0.95, R) >droSec1.super_1 1787455 91 + 14215200 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC-UGACAGACUCGGCGCCA--UCCGUCGUCCCUCCCAAGGAUUCCCC---UUUUUUUCUGA------------- ...........((.....))(((((((..........-.))))))).(((((...--..)))))........((((.....))---)).........------------- ( -15.60, z-score = -0.24, R) >droYak2.chr2L 16792057 90 + 22324452 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC-UGACAGACACGGCGCCA--UCCGCCGACCAUCCCAAGGAUUCCCC---UUUUUU-CUGA------------- ...........((.....))(((((((..........-.))))))).(((((...--..)))))........((((.....))---))....-....------------- ( -19.70, z-score = -1.60, R) >droEre2.scaffold_4929 8148385 90 - 26641161 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC-UGACAGACACGGCGCCA--UCCGUCGUCCCUCCCAAGGAUGCCCC---UUUUUU-CUGA------------- ...........(((((....(((((((..........-.)))))))((((((...--..))))))..........)))))...---......-....------------- ( -19.10, z-score = -0.99, R) >droAna3.scaffold_13266 18034941 109 - 19884421 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC-UGACAGACACGGCGCCACAUCCUGCAUCCCUCCGUUUCCCAAUUCUUCAUCCUUUCUUUUUUUUUCUUCUGA ..................(((..(((....(((((..-((.....)).))))).)))..).))............................................... ( -11.80, z-score = 0.23, R) >dp4.chr3 2126531 95 + 19779522 AAAUAAUAUUAGCAUUAAGUGUUUGUCUCUGUGCCUC-UGACAAACGCGGCGCCG--UCCGUCAUCCCCUCCCUUUUCCCUUUUCCUUUUUUUCAAUA------------ ..................(((((((((..........-.)))))))))((((...--..))))...................................------------ ( -15.40, z-score = -3.02, R) >droPer1.super_2 2297157 95 + 9036312 AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGUGCCUC-UGACAAACGCGGCGCCG--UCCAUCAUCCCCUCCCUUUUCCCUUUUCCUUUUUUUCAAUA------------ ...........((.....(((((((((..........-.))))))))).))....--.........................................------------ ( -15.40, z-score = -2.93, R) >droMoj3.scaffold_6496 8860459 108 - 26866924 AAAUAAUAUUAGCAUUAAGCGUUUGUCUGCGCGCCUCGUGACAGCCGCGGCGCCA--UUCGUCGCCAUCCGUCGCUCCGUUUCUCUAUUUUUUUCUUCUUAAUUUUUUUA (((((.....(((.....((((......)))).....(((((.(..((((((...--..))))))...).)))))...)))....))))).................... ( -20.70, z-score = -0.66, R) >droGri2.scaffold_15245 2473520 75 + 18325388 AAAUAAUAUUAGCAUUAAGCGUUUGUCUGCGCGCCUCGCGACUGCCACGGCGCCA--UCCGUCGUCCUCUGGCUUUU--------------------------------- ...........((.....((((......)))).....))....(((((((((...--..)))))).....)))....--------------------------------- ( -17.10, z-score = 0.51, R) >consensus AAAUAAUAUUAGCAUUAAGCGUUUGUCUCUGCGCCUC_UGACAGACACGGCGCCA__UCCGUCGUCCCUCCCAAGGAUUCCCC___UUUUUUUCUGA_____________ ...........((.....))(((((((............)))))))..((((.......))))............................................... ( -9.99 = -10.26 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:30 2011