
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,054,726 – 4,054,830 |
| Length | 104 |
| Max. P | 0.632622 |

| Location | 4,054,726 – 4,054,830 |
|---|---|
| Length | 104 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.57748 |
| G+C content | 0.57103 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4054726 104 - 21146708 UGCCCGGACGUUUGCGCAGCUUGGCCAUCUUGGCGCGCACAAGGGACACCUUCUCGAUCUCCUUCAUGGCUCGCUCUUUGGCCAGCUGCUCCGGUGUCUUCUUU--------- .(((((((.(....)((((((.(((((....((((.((.((.((((.....))))((......)).)))).))))...)))))))))))))))).)).......--------- ( -40.40, z-score = -2.26, R) >droSim1.chr2R 2737531 104 - 19596830 UGCCCGGACGUUUGCGCAGCUUGGCCAUCUUGGCGCGCACAAGCGACACCUUCUCGAUCUCCUUCAUGGCUCGCUCUUUGGCGAGCUGCUCGGGCGUCUUCUUU--------- .((((((.((((((.((.((...(((.....))))))).))))))......................((((((((....))))))))..)))))).........--------- ( -39.70, z-score = -1.51, R) >droSec1.super_1 1694718 104 - 14215200 UGCCCGGACGCUUGCGCAGCUUGGCCAUCUUGGCGCGCACAAGCGACACCUUCUCGAUCUCCUUCAUGGCUCGCUCUUUGGCGAGCUGCUCGGGCGUCUUCUUU--------- .((((((.((((((.((.((...(((.....))))))).))))))......................((((((((....))))))))..)))))).........--------- ( -42.10, z-score = -2.05, R) >droYak2.chr2L 16704955 104 - 22324452 UGCCUGGACGUUUGCGCAGCUUGGCCAUCUUGGCGCGCACAAGGGACACCUUCUCGAUCUCCUUCAUUGCUCGCUCUUUAGCCAACUGCUCGGGCGUCUUCUUA--------- .....(((((((((.((((.(((((......((((.(((.((((....))))...((......))..))).)))).....))))))))).))))))))).....--------- ( -37.80, z-score = -2.65, R) >droEre2.scaffold_4929 8060945 104 + 26641161 UGCCUGGACGUUUGCGCAGCUUGGCCAUCUUGGCGCGUACAAGGGACACCUUCUCGAUCUCUUUCAUUGCUCGCUCCUUGGCCAGCUGCUCGGGCGUCUUCUUU--------- .....(((((((((.((((((.(((((....((.(((..(((((((.....))))((......)).)))..))).)).))))))))))).))))))))).....--------- ( -45.20, z-score = -4.21, R) >droAna3.scaffold_13266 16360641 104 - 19884421 UGCCGGGCCGUUUCCGCAACUUGGCCAUCUUGGCCCGCACCAGGGACACUUUCUCAAUCUCCUUCAUGGCCCGCUCCUUCGCCAGUUGUUCGGGCGUUUUCUUC--------- .((.(((((((....((.....((((.....)))).))....((((.....))))..........))))))))).....((((.(.....).))))........--------- ( -30.00, z-score = -0.28, R) >dp4.chr3 2033661 104 - 19779522 UCCCAGGGCGCUUGCGGAGCUUGGACAUUUUGGCCCGCACCAGCGACACCUUCUCGAUUUCUUUCAUGGCCCGCUCCUUAGCCAUCUGCUCAGGCGUCUUUUUC--------- ....(((((((((((((((((.(((......(((((((....)))..........((......))..))))...)))..)))..)))))..)))))))))....--------- ( -31.80, z-score = -0.77, R) >droPer1.super_2 2206276 104 - 9036312 UCCCAGGGCGCUUGCGGAGCUUGGACAUUUUGGCCCGCACCAGCGACACCUUCUCGAUUUCUUUCAUGGCCCGCUCCUUAGCCAUCUGCUCAGGCGUCUUUUUC--------- ....(((((((((((((((((.(((......(((((((....)))..........((......))..))))...)))..)))..)))))..)))))))))....--------- ( -31.80, z-score = -0.77, R) >droWil1.scaffold_180701 3532413 104 + 3904529 UGCCUGGACGCUUGCGCAAUUUAGCCAUUUUGGCUCGUACCAUGGACACCUGCUCGAUUUCUUUCAUGGCACGCUCUUUGGCCAGCUGCUCGGGUGUUUUCUUU--------- .(((((..((((.((.(((...((((.....))))(((.(((((((................))))))).)))....))))).))).)..))))).........--------- ( -29.19, z-score = -0.36, R) >droMoj3.scaffold_6496 8745691 104 + 26866924 UGCCAGGUCGUUUCCGUAGCUUGGCCAUUUUGGCGCGCACCAGCGAAACCUUUUCGAUUUCCCUCAUAGCGCGCUCCUUGGCCAUCUGUUCAGGCGUCUUCUUU--------- .(((.((......))(.(((.((((((....(((((((...((.((((.(.....).)))).))....)))))))...))))))...)))).))).........--------- ( -33.70, z-score = -2.02, R) >droGri2.scaffold_15245 2383682 104 - 18325388 UGCCUGGACGCUUGCGUAGCUUGGCCAUUUUGGCGCGUACCAGCGAAACCUUUUCAAUUUCCUUCAUGGCGCGCUCCUUGGCAAGCUGCUCGGGCGUUUUCUUA--------- .....(((((((((.((((((((.(((....((.(((..(((..(((...............))).)))..))).)).))))))))))).))))))))).....--------- ( -41.96, z-score = -2.69, R) >anoGam1.chr3L 25795838 113 + 41284009 GCCCCGGACGGUUGCGUAUCUUGGCCAGCUUGGCCUGCACCAUCGACACCUUGCGCACCUCCUCGAGCGCUCGCUCGCGUGCCAACUGUUUCGCUGUCUUGUUUGCGUCCGCG ....((((((((.(((......((((.....))))))))))..(((.((.(((.((((.....(((((....))))).)))))))..)).))).............))))).. ( -36.00, z-score = 0.21, R) >triCas2.ChLG7 16927411 111 + 17478683 UGUCGCAACUUUUUCGCACUUUUCGUAUCCCACAAGUCACUACCGAAAUUCGUUUUCUUCCGUUUAACGCCAGCUUUCCGACCGACUUCAACGUCCACAACAGCGUUAAUU-- .(((((.........))..................(((......((((.....))))...((.....))..........))).)))...(((((........)))))....-- ( -10.40, z-score = 1.06, R) >consensus UGCCCGGACGUUUGCGCAGCUUGGCCAUCUUGGCGCGCACCAGCGACACCUUCUCGAUCUCCUUCAUGGCUCGCUCCUUGGCCAGCUGCUCGGGCGUCUUCUUU_________ .....(((((((((.((((....(((.....)))......................................((......))...)))).))))))))).............. (-14.64 = -14.72 + 0.09)

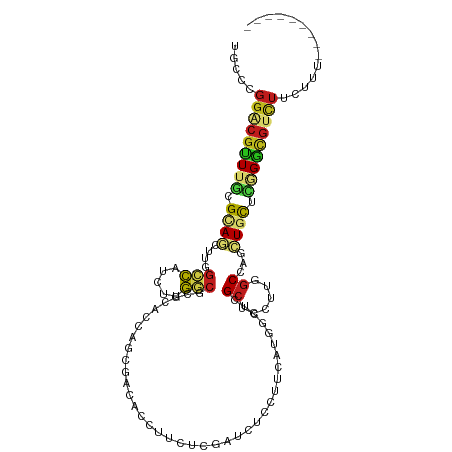
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:22 2011