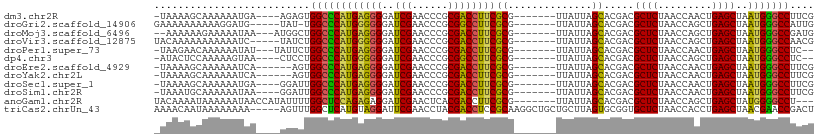
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,045,955 – 4,046,053 |
| Length | 98 |
| Max. P | 0.979040 |

| Location | 4,045,955 – 4,046,053 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.40364 |
| G+C content | 0.50426 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -26.37 |
| Energy contribution | -25.59 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.846915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
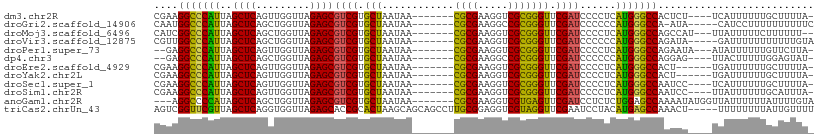
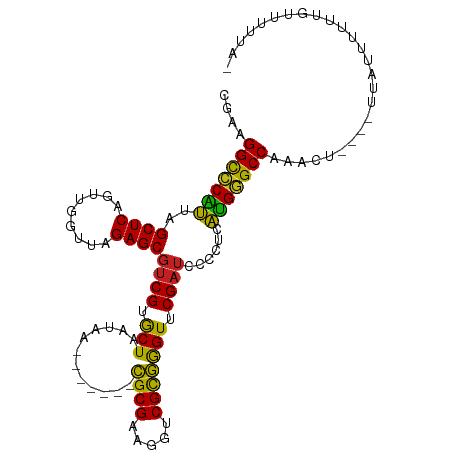
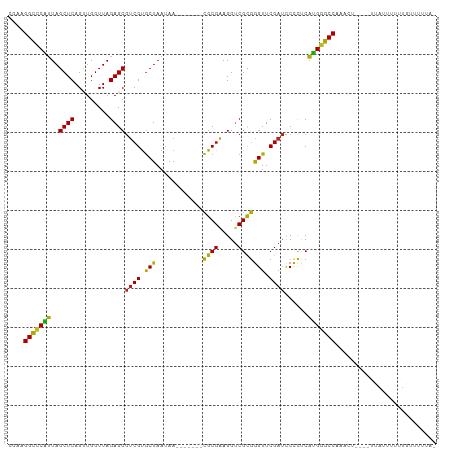
>dm3.chr2R 4045955 98 + 21146708 CGAAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCACUCU----UCAUUUUUUGCUUUUA- ....(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))).....----................- ( -28.40, z-score = -0.63, R) >droGri2.scaffold_14906 10463681 97 - 14172833 CAAUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGCCGCGGGUUCGAUCCCCCCAUGGGCCA-AUA-----CAUCCUUUUUUUUUUC ...((((((((.......((.((((...((((..(....)..)-------)))...))))))(((......)))...))))))))-...-----................ ( -30.80, z-score = -1.49, R) >droMoj3.scaffold_6496 13658311 98 + 26866924 CAUCGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGCCAU---UUAUUUUUCUUUUUU-- ....(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))......---...............-- ( -28.90, z-score = -1.00, R) >droVir3.scaffold_12875 16583227 98 - 20611582 CGUUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCCCAUGGGCCAGAUA-----GAUUUUUUUUUUUGUA ..(((((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))))(((-----((........))))). ( -30.50, z-score = -1.07, R) >droPer1.super_73 77273 97 + 267513 --GAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGAAUA---AUAUUUUUUGUUCUUA- --..(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......)))))))((((((---(......)))))))..- ( -32.70, z-score = -2.30, R) >dp4.chr3 1802856 96 - 19779522 --GAGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGCCGCGGGUUCGAUCCCCCCAUGGGCCAGGAG----UUACUUUUUGGAGUAU- --..(((((((.......((.((((...((((..(....)..)-------)))...))))))(((......)))...))))))).....----.(((((....))))).- ( -32.20, z-score = -0.36, R) >droEre2.scaffold_4929 8051700 96 - 26641161 CGAAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCACU------UGAUUUUUUGCUUUUA- ....(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......)))))))...------................- ( -28.40, z-score = -0.42, R) >droYak2.chr2L 16695602 96 + 22324452 CGAAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCACU------UGAUUUUUUGCUUUUA- ....(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......)))))))...------................- ( -28.40, z-score = -0.42, R) >droSec1.super_1 1686025 98 + 14215200 CGAAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAAUCC----UCAUUUUUUGCUUUUA- ....(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))).....----................- ( -28.40, z-score = -0.61, R) >droSim1.chr2R 2728304 98 + 19596830 CGAAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAAUCC----UUAUUUUUUGCAUUUA- ....(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))).....----................- ( -28.40, z-score = -0.72, R) >anoGam1.chr2R 13947731 100 - 62725911 ---AGGCCCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCCAAAAUAUGGUUAUUUUUUAUUUUGUA ---.(((.(((((((...))))...((((.((((.(((....(-------(((....).))).))).)))).)))).))).)))((((((....)))))).......... ( -26.40, z-score = -0.66, R) >triCas2.chrUn_43 98014 105 - 275627 AGUCGGUUCGUUAGCUCAGGUGGUUAGAGCACCGCACUAAGCAGCAGCCUUGCGGAGGUCGUAGGUUCGAAUCCUACAUGAGCCAAACU-----UUUUUUUUAUUGUUUU ....((((((((.(((.(((((((......))))).)).)))))).(((((...))))).(((((.......)))))..))))).....-----................ ( -29.80, z-score = -1.31, R) >consensus CGAAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA_______CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAAACU____UUAUUUUUUGUUUUUA_ ....(((((((..((((.........))))((((.(((............((((.....))))))).))))......))))))).......................... (-26.37 = -25.59 + -0.78)
| Location | 4,045,955 – 4,046,053 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.40364 |
| G+C content | 0.50426 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -25.42 |
| Energy contribution | -24.94 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
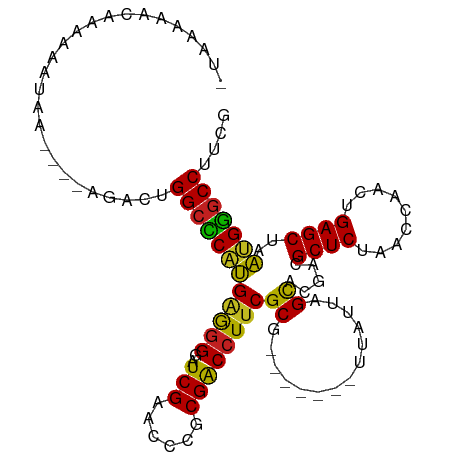
>dm3.chr2R 4045955 98 - 21146708 -UAAAAGCAAAAAAUGA----AGAGUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUUCG -................----.(((.(((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))))). ( -29.50, z-score = -2.00, R) >droGri2.scaffold_14906 10463681 97 + 14172833 GAAAAAAAAAAGGAUG-----UAU-UGGCCCAUGGGGGGAUCGAACCCGCGGCCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCAUUG ................-----...-(((((((((.(((.......))).).....(((.(-------(.....)).))).((((.........))))..))))))))... ( -28.80, z-score = -0.40, R) >droMoj3.scaffold_6496 13658311 98 - 26866924 --AAAAAAGAAAAAUAA---AUGGCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGAUG --...............---.....((((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))))... ( -28.30, z-score = -1.38, R) >droVir3.scaffold_12875 16583227 98 + 20611582 UACAAAAAAAAAAAUC-----UAUCUGGCCCAUGGGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCAACG ................-----....((((((((.(((((.(((......))))))))((.-------......)).....((((.........))))..))))))))... ( -29.40, z-score = -2.22, R) >droPer1.super_73 77273 97 - 267513 -UAAGAACAAAAAAUAU---UAUUCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUC-- -................---......(((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))..-- ( -27.50, z-score = -2.34, R) >dp4.chr3 1802856 96 + 19779522 -AUACUCCAAAAAGUAA----CUCCUGGCCCAUGGGGGGAUCGAACCCGCGGCCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCUC-- -.((((......)))).----.....((((((((.(((.......))).).....(((.(-------(.....)).))).((((.........))))..)))))))..-- ( -28.60, z-score = -0.17, R) >droEre2.scaffold_4929 8051700 96 + 26641161 -UAAAAGCAAAAAAUCA------AGUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUUCG -................------...(((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))).... ( -27.50, z-score = -1.65, R) >droYak2.chr2L 16695602 96 - 22324452 -UAAAAGCAAAAAAUCA------AGUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUUCG -................------...(((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))).... ( -27.50, z-score = -1.65, R) >droSec1.super_1 1686025 98 - 14215200 -UAAAAGCAAAAAAUGA----GGAUUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUUCG -................----.((..(((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))).)). ( -28.40, z-score = -1.57, R) >droSim1.chr2R 2728304 98 - 19596830 -UAAAUGCAAAAAAUAA----GGAUUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUUCG -................----.((..(((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))).)). ( -28.40, z-score = -1.78, R) >anoGam1.chr2R 13947731 100 + 62725911 UACAAAAUAAAAAAUAACCAUAUUUUGGCUCCAGAGAGGAUCGAACUCACGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGGGCCU--- ..........((((((....))))))(((((((..((((.(((......))).))))((.-------......)).....((((.........))))..))))))).--- ( -25.90, z-score = -2.27, R) >triCas2.chrUn_43 98014 105 + 275627 AAAACAAUAAAAAAAA-----AGUUUGGCUCAUGUAGGAUUCGAACCUACGACCUCCGCAAGGCUGCUGCUUAGUGCGGUGCUCUAACCACCUGAGCUAACGAACCGACU ................-----.(((.((((((.(((((.......)))))((.(.(((((.(((....)))...))))).).))........)))))))))......... ( -28.90, z-score = -2.59, R) >consensus _UAAAAACAAAAAAUAA____AGACUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG_______UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUUCG ..........................(((((((..(((.......)))(((.....))).....................((((.........))))..))))))).... (-25.42 = -24.94 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:21 2011