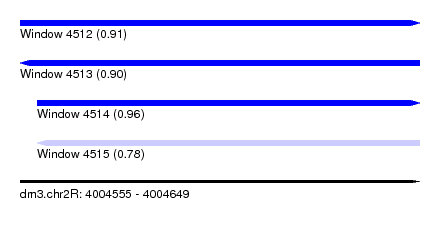
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,004,555 – 4,004,649 |
| Length | 94 |
| Max. P | 0.960956 |

| Location | 4,004,555 – 4,004,649 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 63.45 |
| Shannon entropy | 0.76079 |
| G+C content | 0.48982 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -9.86 |
| Energy contribution | -9.61 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
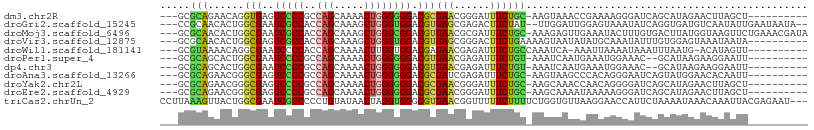
>dm3.chr2R 4004555 94 + 21146708 ---GCGCAGAACAGGUGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAACGGGAUUUCUGC-AAGUAAACCGAAAAGGGAUCAGCAUAGAACUUAGCU---------- ---(((((((((..((..((((((.((((.....))))))))))....))..)..))))))-((((...((.....))..((......)))))).)).---------- ( -31.60, z-score = -2.06, R) >droGri2.scaffold_15245 14917295 101 + 18325388 ---CCGCAACACUGGCGAAUCGCCACCAGCAAAGCUGGGUGGACGUGAGCGAGACUUCUAU--UUGGAUUGGAGUAAAUAUCAGGUGAUGUCAAUAUUGAAUAAUA-- ---((((.....(((((...)))))(((((...)))))))))((.(((.....(((((...--.......))))).....))).))....................-- ( -27.20, z-score = -1.07, R) >droMoj3.scaffold_6496 17913601 104 - 26866924 ---GCGCAACACUGGCGAAUCGCCACCAGCAAAGCUGGGCGGACGUGAACGCGAUUUCUGC-AAAGAGUUGAAAUACUUUGUGACUUAUGGUAAGUUCUGAAACGAUA ---.(((.....(((((...)))))(((((...))))))))..((....))((.((((.((-((((.((....)).))))))((((((...))))))..))))))... ( -26.30, z-score = 0.00, R) >droVir3.scaffold_12875 14895178 95 + 20611582 ---GCGCAACACUGGCGAGUCGCCACCAGCAAAGCUGGGUGGACGUGAGCGGGACUUCUGAAAAGUAAUAUAUGCAAAUAUUUGUGGAGUAAAUAAUA---------- ---.(((((((((((((...)))))(((((...)))))......))).(((......((....)).......)))......)))))............---------- ( -23.32, z-score = -0.16, R) >droWil1.scaffold_181141 3070394 93 - 5303230 ---GCGUAAAACAGGCGAAUCCCCACCAGCAAAACUUGGUGGACGAGAACGAGAUUUCUGCCAAAUCA-AAAUUAAAAUAAAUUUAAUG-ACAUAGUU---------- ---..((......((((((((.(((((((......))))))).((....)).)))))..)))......-..((((((.....)))))).-))......---------- ( -17.20, z-score = -1.50, R) >droPer1.super_4 5448279 92 - 7162766 ---GCGCAGCACUGGCGAAUCCCCGCCAGCAAAACUGGGGGGACGUGAACGAGAUUUCUGU-AAAUCAAUGAAAUGGAAAC--GCAUAAGAAGGAAUU---------- ---((((((..........(((((.((((.....)))))))))((....))......))))-.............(....)--)).............---------- ( -24.00, z-score = -0.98, R) >dp4.chr3 11975476 92 - 19779522 ---GCGCAGCACUGGCGAAUCCCCGCCAGCAAAACUGGGGGGACGUGAACGAGAUUUCUGU-AAAUCAAUGAAAUGGAAAC--GCAUAAGAAGGAAUU---------- ---((((((..........(((((.((((.....)))))))))((....))......))))-.............(....)--)).............---------- ( -24.00, z-score = -0.98, R) >droAna3.scaffold_13266 16317704 94 + 19884421 ---GCGCAGAACGGGCGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAUCGAGAUUUCUGC-AAGUAAGCCCACAGGGAAUCAGUAUGGAACACAAUU---------- ---..((((((((..((.((((((.((((.....)))))))))).))..))....))))))-.......(((...)))....................---------- ( -33.80, z-score = -2.49, R) >droYak2.chr2L 16653869 94 + 22324452 ---GCGCAGAACGGGCGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAACGGGAUUUCUGC-AAGCAAACCAACAGGGGAUCAGCAUAGAACUUAGCU---------- ---((((((((((..((.((((((.((((.....)))))))))).))..))....))))))-..((...((.....)).....))..........)).---------- ( -36.70, z-score = -3.15, R) >droEre2.scaffold_4929 8010937 94 - 26641161 ---GCGCAGAACGGGCGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAACGGGAUUUCUGC-AAGCAAAAUAAAAAGGGAUCAGCAUAGAACUUAGCU---------- ---((((((((((..((.((((((.((((.....)))))))))).))..))....))))))-..............(((.((......)).))).)).---------- ( -35.00, z-score = -3.28, R) >triCas2.chrUn_2 561200 105 + 1160328 CCUUAAAGUUACUGGCGAAUCGCCCCCUGUAUAACUAGGUGGGCGUGAACGGUUUUUCUUUUCUGGUGUUAAGGAACCAUUCUAAAAUAAACAAAUUACGAGAAU--- ((((((...(((..(.(((.(((((((((......)))).))))).(((......)))))).)..)))))))))....(((((..(((......)))...)))))--- ( -23.50, z-score = -0.47, R) >consensus ___GCGCAGAACUGGCGAAUCCCCGCCAGCAAAACUGGGGGGACGUGAACGAGAUUUCUGC_AAGUAAAUGAAAAAGAAAUCAGCAUAGAACAUAAUU__________ ..............(((..(((((..(((.....)))))))).))).............................................................. ( -9.86 = -9.61 + -0.25)
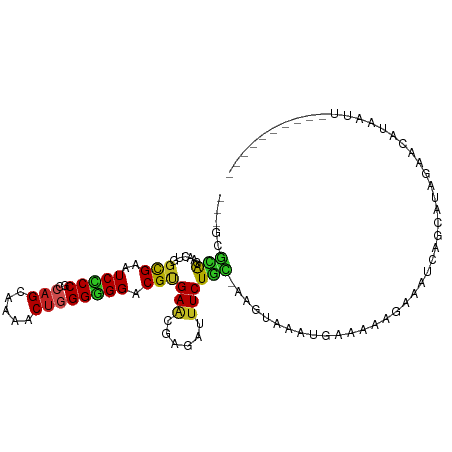
| Location | 4,004,555 – 4,004,649 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 63.45 |
| Shannon entropy | 0.76079 |
| G+C content | 0.48982 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -7.98 |
| Energy contribution | -7.67 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.902830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4004555 94 - 21146708 ----------AGCUAAGUUCUAUGCUGAUCCCUUUUCGGUUUACUU-GCAGAAAUCCCGUUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCACCUGUUCUGCGC--- ----------.((.((((.....(((((.......)))))..))))-((((((.....((..(.((((((((((.....)))).)))))).)))...))))))))--- ( -32.70, z-score = -3.20, R) >droGri2.scaffold_15245 14917295 101 - 18325388 --UAUUAUUCAAUAUUGACAUCACCUGAUAUUUACUCCAAUCCAA--AUAGAAGUCUCGCUCACGUCCACCCAGCUUUGCUGGUGGCGAUUCGCCAGUGUUGCGG--- --..............((((((....)))...........((...--...)).))).(((.(((......(((((...)))))(((((...))))))))..))).--- ( -21.60, z-score = -0.87, R) >droMoj3.scaffold_6496 17913601 104 + 26866924 UAUCGUUUCAGAACUUACCAUAAGUCACAAAGUAUUUCAACUCUUU-GCAGAAAUCGCGUUCACGUCCGCCCAGCUUUGCUGGUGGCGAUUCGCCAGUGUUGCGC--- ....(((((.((.((((...)))))).(((((..........))))-)..))))).((((.(((......(((((...)))))(((((...))))))))..))))--- ( -25.80, z-score = -0.99, R) >droVir3.scaffold_12875 14895178 95 - 20611582 ----------UAUUAUUUACUCCACAAAUAUUUGCAUAUAUUACUUUUCAGAAGUCCCGCUCACGUCCACCCAGCUUUGCUGGUGGCGACUCGCCAGUGUUGCGC--- ----------................(((((......)))))(((((...)))))..(((.(((......(((((...)))))(((((...))))))))..))).--- ( -19.40, z-score = -0.83, R) >droWil1.scaffold_181141 3070394 93 + 5303230 ----------AACUAUGU-CAUUAAAUUUAUUUUAAUUU-UGAUUUGGCAGAAAUCUCGUUCUCGUCCACCAAGUUUUGCUGGUGGGGAUUCGCCUGUUUUACGC--- ----------......((-((..(((((......)))))-))))..(((((((......))))..(((((((........))))))).....)))..........--- ( -16.50, z-score = -0.07, R) >droPer1.super_4 5448279 92 + 7162766 ----------AAUUCCUUCUUAUGC--GUUUCCAUUUCAUUGAUUU-ACAGAAAUCUCGUUCACGUCCCCCCAGUUUUGCUGGCGGGGAUUCGCCAGUGCUGCGC--- ----------.............((--((...((((.....(((((-....)))))........((((((((((.....)))).)))))).....))))..))))--- ( -24.10, z-score = -1.71, R) >dp4.chr3 11975476 92 + 19779522 ----------AAUUCCUUCUUAUGC--GUUUCCAUUUCAUUGAUUU-ACAGAAAUCUCGUUCACGUCCCCCCAGUUUUGCUGGCGGGGAUUCGCCAGUGCUGCGC--- ----------.............((--((...((((.....(((((-....)))))........((((((((((.....)))).)))))).....))))..))))--- ( -24.10, z-score = -1.71, R) >droAna3.scaffold_13266 16317704 94 - 19884421 ----------AAUUGUGUUCCAUACUGAUUCCCUGUGGGCUUACUU-GCAGAAAUCUCGAUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCGCCCGUUCUGCGC--- ----------....(((.((((((.........))))))..)))..-((((((....((..((.((((((((((.....)))).)))))).))..))))))))..--- ( -35.10, z-score = -3.08, R) >droYak2.chr2L 16653869 94 - 22324452 ----------AGCUAAGUUCUAUGCUGAUCCCCUGUUGGUUUGCUU-GCAGAAAUCCCGUUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCGCCCGUUCUGCGC--- ----------.((.((((.....((..((.....))..))..))))-((((((....((..((.((((((((((.....)))).)))))).))..))))))))))--- ( -35.90, z-score = -3.33, R) >droEre2.scaffold_4929 8010937 94 + 26641161 ----------AGCUAAGUUCUAUGCUGAUCCCUUUUUAUUUUGCUU-GCAGAAAUCCCGUUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCGCCCGUUCUGCGC--- ----------(((..........)))....................-((((((....((..((.((((((((((.....)))).)))))).))..))))))))..--- ( -32.60, z-score = -3.38, R) >triCas2.chrUn_2 561200 105 - 1160328 ---AUUCUCGUAAUUUGUUUAUUUUAGAAUGGUUCCUUAACACCAGAAAAGAAAAACCGUUCACGCCCACCUAGUUAUACAGGGGGCGAUUCGCCAGUAACUUUAAGG ---......((.(((.((........((((((((.(((..........)))...)))))))).(((((.(((........))))))))....)).))).))....... ( -22.30, z-score = -0.85, R) >consensus __________AAUUAUGUUCUAUGCUGAUAUCUUUUUCAUUUACUU_GCAGAAAUCCCGUUCACGUCCCCCCAGUUUUGCUGGCGGGGAUUCGCCAGUGCUGCGC___ .........................................................................((...((.(((((....))))).))...))..... ( -7.98 = -7.67 + -0.30)
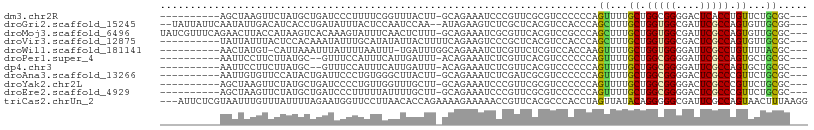
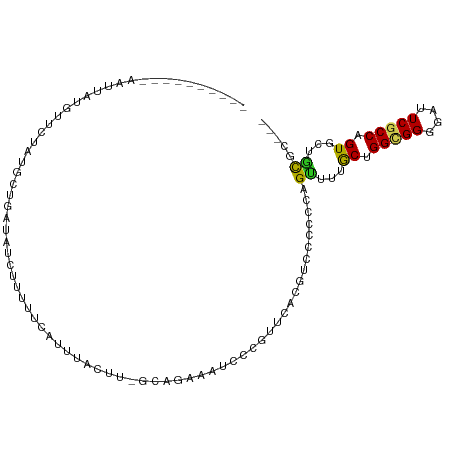
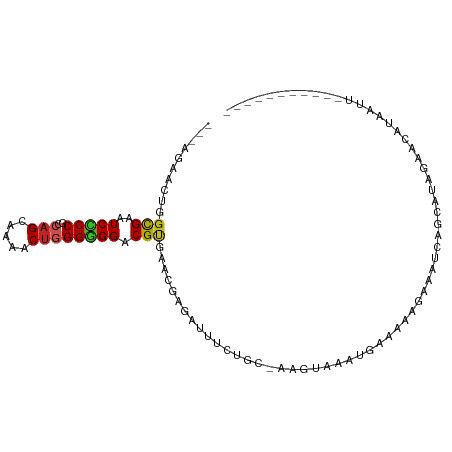
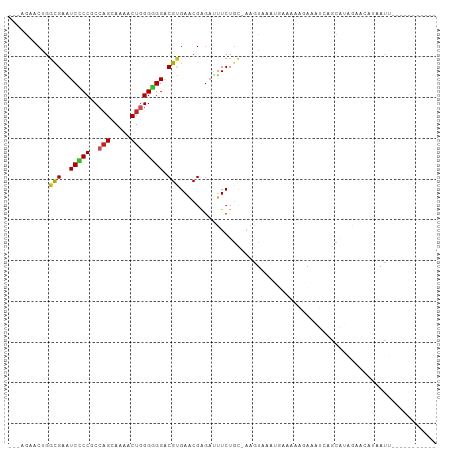
| Location | 4,004,559 – 4,004,649 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 62.26 |
| Shannon entropy | 0.77767 |
| G+C content | 0.46913 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -9.86 |
| Energy contribution | -9.61 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4004559 90 + 21146708 ---AGAACAGGUGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAACGGGAUUUCUGC-AAGUAAACCGAAAAGGGAUCAGCAUAGAACUUAGCU----------- ---.....((((..((((((.((((.....))))))))))((......(((..((..-..(.....)....))..))).)).....))))....----------- ( -26.80, z-score = -1.23, R) >droGri2.scaffold_15245 14917299 97 + 18325388 ---AACACUGGCGAAUCGCCACCAGCAAAGCUGGGUGGACGUGAGCGAGACUUCUAUUUGGAU-UGGAGUAAAUAUCAGGUGAUGUCAAUAUUG-AAUAAUA--- ---..((((((((...)))))(((((...))))))))(((((.(.(((.(((((.........-.))))).....)).).).))))).......-.......--- ( -26.20, z-score = -1.50, R) >droMoj3.scaffold_6496 17913605 100 - 26866924 ---AACACUGGCGAAUCGCCACCAGCAAAGCUGGGCGGACGUGAACGCGAUUUCUGC-AAAGAGUUGAAAUACUUUGUGACUUAUGGUAAGUUCUGAAACGAUA- ---..((((((((...)))))(((((...)))))((((((((....)))...)))))-..(((((......))))))))(((((...)))))............- ( -24.50, z-score = -0.30, R) >droVir3.scaffold_12875 14895182 91 + 20611582 ---AACACUGGCGAGUCGCCACCAGCAAAGCUGGGUGGACGUGAGCGGGACUUCUGAAAAGUAAUAUAUGCAAAUAUUUGUGGAGUAAAUAAUA----------- ---....((.((....((((((((((...))).))))).))...)).))(((((....(((((...........)))))..)))))........----------- ( -21.90, z-score = -0.66, R) >droWil1.scaffold_181141 3070398 89 - 5303230 ---AAAACAGGCGAAUCCCCACCAGCAAAACUUGGUGGACGAGAACGAGAUUUCUGCCAAAUCA-AAAUUAAAAUAAAUUUAAUG-ACAUAGUU----------- ---..(((.((((((((.(((((((......))))))).((....)).)))))..)))......-..((((((.....)))))).-.....)))----------- ( -17.20, z-score = -2.09, R) >droPer1.super_4 5448283 88 - 7162766 ---AGCACUGGCGAAUCCCCGCCAGCAAAACUGGGGGGACGUGAACGAGAUUUCUGU-AAAUCAAUGAAAUGGAAAC--GCAUAAGAAGGAAUU----------- ---....((.(((..(((((.((((.....)))))))))((....)).(((((....-))))).............)--))...))........----------- ( -23.40, z-score = -1.56, R) >dp4.chr3 11975480 88 - 19779522 ---AGCACUGGCGAAUCCCCGCCAGCAAAACUGGGGGGACGUGAACGAGAUUUCUGU-AAAUCAAUGAAAUGGAAAC--GCAUAAGAAGGAAUU----------- ---....((.(((..(((((.((((.....)))))))))((....)).(((((....-))))).............)--))...))........----------- ( -23.40, z-score = -1.56, R) >droAna3.scaffold_13266 16317708 90 + 19884421 ---AGAACGGGCGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAUCGAGAUUUCUGC-AAGUAAGCCCACAGGGAAUCAGUAUGGAACACAAUU----------- ---....((..((.((((((.((((.....)))))))))).))..)).(((((((((-......)).....)))))))................----------- ( -30.60, z-score = -2.12, R) >droYak2.chr2L 16653873 90 + 22324452 ---AGAACGGGCGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAACGGGAUUUCUGC-AAGCAAACCAACAGGGGAUCAGCAUAGAACUUAGCU----------- ---....((..((.((((((.((((.....)))))))))).))..)).(((..((..-.............))..)))(((..........)))----------- ( -30.56, z-score = -1.94, R) >droEre2.scaffold_4929 8010941 90 - 26641161 ---AGAACGGGCGAGUCCCCGCCAGCAAAACUGGGGGGACGCGAACGGGAUUUCUGC-AAGCAAAAUAAAAAGGGAUCAGCAUAGAACUUAGCU----------- ---....((..((.((((((.((((.....)))))))))).))..)).(((..((..-.............))..)))(((..........)))----------- ( -31.16, z-score = -2.82, R) >triCas2.chrUn_2 561204 105 + 1160328 AAAGUUACUGGCGAAUCGCCCCCUGUAUAACUAGGUGGGCGUGAACGGUUUUUCUUUUCUGGUGUUAAGGAACCAUUCUAAAAUAAACAAAUUACGAGAAUAUUU ((((..((((......(((((((((......)))).)))))....))))....))))...(((........)))(((((..(((......)))...))))).... ( -22.90, z-score = -0.45, R) >consensus ___AGAACUGGCGAAUCCCCGCCAGCAAAACUGGGGGGACGUGAACGAGAUUUCUGC_AAGUAAAUGAAAAAGAAAUCAGCAUAGAACAUAAUU___________ ..........(((..(((((..(((.....)))))))).)))............................................................... ( -9.86 = -9.61 + -0.25)
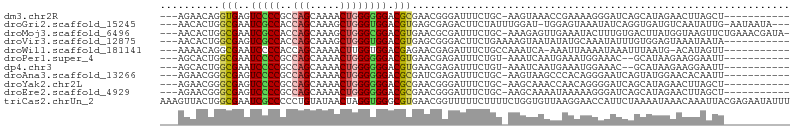
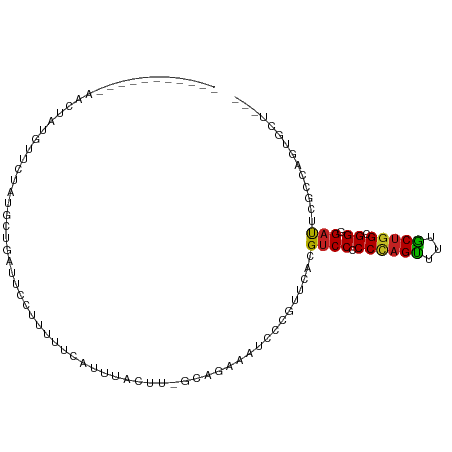
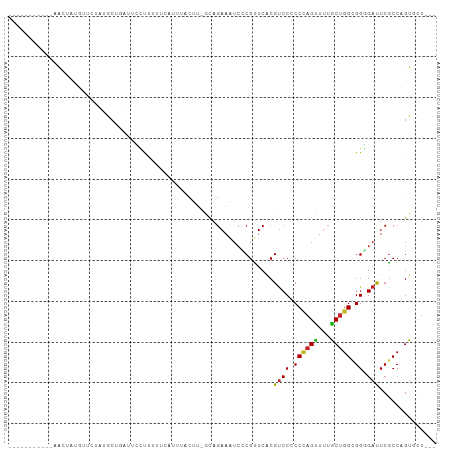
| Location | 4,004,559 – 4,004,649 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 62.26 |
| Shannon entropy | 0.77767 |
| G+C content | 0.46913 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -6.45 |
| Energy contribution | -6.12 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4004559 90 - 21146708 -----------AGCUAAGUUCUAUGCUGAUCCCUUUUCGGUUUACUU-GCAGAAAUCCCGUUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCACCUGUUCU--- -----------.((.((((.....(((((.......)))))..))))-)).(((......)))(.((((((((((.....)))).)))))).).........--- ( -28.90, z-score = -2.85, R) >droGri2.scaffold_15245 14917299 97 - 18325388 ---UAUUAUU-CAAUAUUGACAUCACCUGAUAUUUACUCCA-AUCCAAAUAGAAGUCUCGCUCACGUCCACCCAGCUUUGCUGGUGGCGAUUCGCCAGUGUU--- ---.......-.(((((((.(.....((..(((((......-....)))))..)).........((.(((((.(((...))))))))))....).)))))))--- ( -17.30, z-score = -0.45, R) >droMoj3.scaffold_6496 17913605 100 + 26866924 -UAUCGUUUCAGAACUUACCAUAAGUCACAAAGUAUUUCAACUCUUU-GCAGAAAUCGCGUUCACGUCCGCCCAGCUUUGCUGGUGGCGAUUCGCCAGUGUU--- -....(((((.((.((((...)))))).(((((..........))))-)..))))).(((....((.(((((.(((...))))))))))...))).......--- ( -21.00, z-score = -0.43, R) >droVir3.scaffold_12875 14895182 91 - 20611582 -----------UAUUAUUUACUCCACAAAUAUUUGCAUAUAUUACUUUUCAGAAGUCCCGCUCACGUCCACCCAGCUUUGCUGGUGGCGACUCGCCAGUGUU--- -----------................((((((.((.......(((((...)))))........((.(((((.(((...))))))))))....)).))))))--- ( -17.50, z-score = -1.19, R) >droWil1.scaffold_181141 3070398 89 + 5303230 -----------AACUAUGU-CAUUAAAUUUAUUUUAAUUU-UGAUUUGGCAGAAAUCUCGUUCUCGUCCACCAAGUUUUGCUGGUGGGGAUUCGCCUGUUUU--- -----------......((-((..(((((......)))))-))))..(((((((......))))..(((((((........))))))).....)))......--- ( -16.50, z-score = -0.46, R) >droPer1.super_4 5448283 88 + 7162766 -----------AAUUCCUUCUUAUGC--GUUUCCAUUUCAUUGAUUU-ACAGAAAUCUCGUUCACGUCCCCCCAGUUUUGCUGGCGGGGAUUCGCCAGUGCU--- -----------.............((--(.............(((((-....)))))........((((((((((.....)))).)))))).))).......--- ( -21.40, z-score = -1.73, R) >dp4.chr3 11975480 88 + 19779522 -----------AAUUCCUUCUUAUGC--GUUUCCAUUUCAUUGAUUU-ACAGAAAUCUCGUUCACGUCCCCCCAGUUUUGCUGGCGGGGAUUCGCCAGUGCU--- -----------.............((--(.............(((((-....)))))........((((((((((.....)))).)))))).))).......--- ( -21.40, z-score = -1.73, R) >droAna3.scaffold_13266 16317708 90 - 19884421 -----------AAUUGUGUUCCAUACUGAUUCCCUGUGGGCUUACUU-GCAGAAAUCUCGAUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCGCCCGUUCU--- -----------................((((..(((..((....)).-.))).)))).((..((.((((((((((.....)))).)))))).))..))....--- ( -31.40, z-score = -2.78, R) >droYak2.chr2L 16653873 90 - 22324452 -----------AGCUAAGUUCUAUGCUGAUCCCCUGUUGGUUUGCUU-GCAGAAAUCCCGUUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCGCCCGUUCU--- -----------.((.((((.....((..((.....))..))..))))-))((((....((..((.((((((((((.....)))).)))))).))..))))))--- ( -31.20, z-score = -2.71, R) >droEre2.scaffold_4929 8010941 90 + 26641161 -----------AGCUAAGUUCUAUGCUGAUCCCUUUUUAUUUUGCUU-GCAGAAAUCCCGUUCGCGUCCCCCCAGUUUUGCUGGCGGGGACUCGCCCGUUCU--- -----------.((.((((...(((..((....))..)))...))))-))((((....((..((.((((((((((.....)))).)))))).))..))))))--- ( -27.60, z-score = -2.63, R) >triCas2.chrUn_2 561204 105 - 1160328 AAAUAUUCUCGUAAUUUGUUUAUUUUAGAAUGGUUCCUUAACACCAGAAAAGAAAAACCGUUCACGCCCACCUAGUUAUACAGGGGGCGAUUCGCCAGUAACUUU ..........((.(((.((........((((((((.(((..........)))...)))))))).(((((.(((........))))))))....)).))).))... ( -22.30, z-score = -1.35, R) >consensus ___________AACUAUGUUCUAUGCUGAUUCCUUUUUCAUUUACUU_GCAGAAAUCCCGUUCACGUCCCCCCAGUUUUGCUGGCGGGGAUUCGCCAGUGCU___ .................................................................((((.((((((...))))).)).))).............. ( -6.45 = -6.12 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:15 2011