
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,973,074 – 3,973,179 |
| Length | 105 |
| Max. P | 0.604545 |

| Location | 3,973,074 – 3,973,169 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.05 |
| Shannon entropy | 0.70633 |
| G+C content | 0.49431 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.61 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
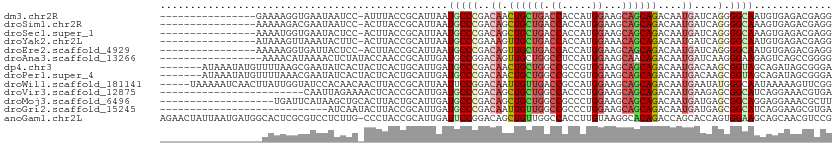
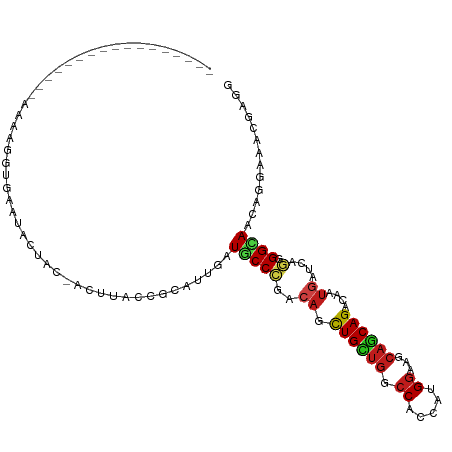
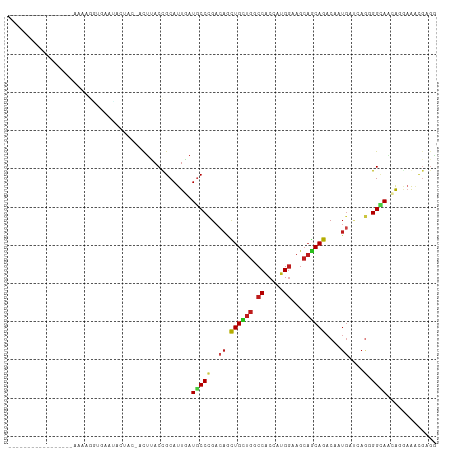
>dm3.chr2R 3973074 95 - 21146708 ----------------GAAAAGGUGAAUAAUCC-AUUUACCGCAUUAAUGCCCGACAACUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAUGUGAGACGAGG ----------------.....(((((((.....-)))))))((((...(((((.....((((((.((.....))...)))))).(........))))))))))......... ( -26.60, z-score = -2.25, R) >droSim1.chr2R 2672563 95 - 19596830 ----------------AAAAAGACGAAUAAUCC-ACUUACCGCAUUAAUGCCCGACAGCUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAAGUGAGACGAGG ----------------.................-.(((..(.((((..(((((....((((((..((.....)).)))))).((......))..))))).)))).)..))). ( -24.60, z-score = -2.22, R) >droSec1.super_1 1617283 95 - 14215200 ----------------AAAAUGGUGAAUACUCC-ACUUACCGCAUUAAUGCCCGACAGCUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAAGUGAGACGAGG ----------------....(((........))-)(((..(.((((..(((((....((((((..((.....)).)))))).((......))..))))).)))).)..))). ( -26.20, z-score = -1.56, R) >droYak2.chr2L 16630245 95 - 22324452 ----------------AUAAAGUUAAAUACUUC-ACUUACCGCAUUAAUGCCCGAAAGUUGCUGACCACCAUGGAAACAGCAGACAAUGAUCAGGGGCAAUGUGAGACGAGG ----------------.............((((-......(((((...(((((((..(((((((.......((....)).))).))))..))..))))))))))....)))) ( -21.30, z-score = -1.15, R) >droEre2.scaffold_4929 7986285 95 + 26641161 ----------------AAAAAGGUGAUUACUCC-ACUUACCGCAUUAAUGCCCGACAGUUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAUGUGAGACGAGG ----------------....(((((.......)-))))..(((((...(((((....((((((..((.....)).)))))).((......))..))))))))))........ ( -25.90, z-score = -1.42, R) >droAna3.scaffold_13266 16289543 95 - 19884421 -----------------AAAACAUAAAACUCUAUACCAACCGCAUUGAUGCCGGACAGUUGCUGGCCUCCAUGGAAGCAACAGACAAUGAUCAAGGGAAGAGUCAGCCGGGG -----------------..........(((((...((..(((((....)).)))...((((((..((.....)).)))))).............))..)))))......... ( -22.20, z-score = -0.06, R) >dp4.chr3 11950112 105 + 19779522 -------AUAAAUAUGUUUUAAGCGAAUAUCACUACUCACUGCAUUGAUGCCCGACAACUGCUGGCCGCCGUGGAAGCAGCAGACAAUGACAAGCGGUAGCAGAUAGCGGGA -------....................(((((...(.....)...)))))((((....((((((.((((..((....((........)).)).))))))))))....)))). ( -24.40, z-score = 0.80, R) >droPer1.super_4 5422682 105 + 7162766 -------AUAAAUAUGUUUUAAACGAAUAUCACUACUCACUGCAUUGAUGCCCGACAACUGCUGGCCGCCGUGGAAGCAGCAGACAAUGACAAGCGGUAGCAGAUAGCGGGA -------....................(((((...(.....)...)))))((((....((((((.((((..((....((........)).)).))))))))))....)))). ( -24.40, z-score = 0.25, R) >droWil1.scaffold_181141 3002236 107 + 5303230 -----UAAAAAUCAACUUAUUGGUAUCCACAACAACUUACCGCAUUAAUUCCGGACAAUUGUUGACCGCCAUGGAAGCAGCAGACAAUGAAUAUGGGCAAUAAAAAGUUCGG -----................((((............)))).........((((((..((((((....(((((....((........))..))))).))))))...)))))) ( -18.40, z-score = 0.04, R) >droVir3.scaffold_12875 14869856 88 - 20611582 ------------------------CAAUUAGAAAACUCACCGCAUUGAUGCCCGACAGCUGCUGGCCACCCUGGAAGCAGCAGACAAUGAAGAGCGGCAUCAGGAAACGUGA ------------------------............((((.....((((((((....((((((..((.....)).))))))...(......).).)))))))(....))))) ( -27.20, z-score = -2.02, R) >droMoj3.scaffold_6496 19842936 93 + 26866924 -------------------UGAUUCAUAAGCUGCACUUACUGCAUUGAUGCCCGACAGCUGCUGGCCGCCCUGGAAGCAGCAGACAAUGAUGAGCGGCAGGAGGAAACGCUU -------------------..........(((((.(......(((((.((.....))((((((..((.....)).))))))...)))))..).)))))(((.(....).))) ( -25.70, z-score = 0.62, R) >droGri2.scaffold_15245 14891174 84 - 18325388 ----------------------------AUCAAUACUUACCGCAUUGAUGCCCGACAAUUGUUGGCCGCCCUGGAAGCAGCAGACAAUGAUGAGCGGCAUCAGGAAGCGUGA ----------------------------...........((((.(((((((((....((((((.((.((.......)).)).)))))).....).))))))))...))).). ( -25.70, z-score = -1.20, R) >anoGam1.chr2L 12132598 111 - 48795086 AGAACUAUUAAUGAUGGCACUCGCGUCCUCUUG-CCCUACCGCAUUGAUUCCGGACAGCUGUUGGCCACCUUGUAAGGCACAGACCAGCACCAGUGGAAGCAGCAACGUCCG ......(((((((.(((.....(((......))-)....))))))))))..(((((.((((((..((((..(((..((......)).)))...)))).))))))...))))) ( -30.70, z-score = -0.24, R) >consensus _________________AAAAGGUGAAUACUAC_ACUUACCGCAUUGAUGCCCGACAGCUGCUGGCCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAACAGGAAACGAGG ................................................(((((..((.((((((.((.....))...))))))....))....).))))............. (-10.55 = -10.61 + 0.06)
| Location | 3,973,084 – 3,973,179 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 65.12 |
| Shannon entropy | 0.76676 |
| G+C content | 0.47793 |
| Mean single sequence MFE | -21.71 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.61 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
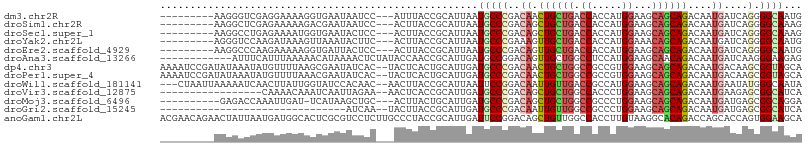
>dm3.chr2R 3973084 95 - 21146708 ---------AAGGGUCGAGGAAAAGGUGAAUAAUCC---AUUUACCGCAUUAAUGCCCGACAACUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAUG ---------...(((((.((....(((((((.....---)))))))(((....))))).....((((((.((.....))...))))))....))))).......... ( -25.40, z-score = -1.18, R) >droSim1.chr2R 2672573 95 - 19596830 ---------AAGGCUCGAGAAAAAGACGAAUAAUCC---ACUUACCGCAUUAAUGCCCGACAGCUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAAG ---------...........................---..............(((((....((((((..((.....)).)))))).((......))..)))))... ( -21.50, z-score = -0.89, R) >droSec1.super_1 1617293 95 - 14215200 ---------AAGGCCUGAGAAAAUGGUGAAUACUCC---ACUUACCGCAUUAAUGCCCGACAGCUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAAG ---------...............(((((.......---..))))).......(((((....((((((..((.....)).)))))).((......))..)))))... ( -25.10, z-score = -0.95, R) >droYak2.chr2L 16630255 95 - 22324452 ---------AGGGUCCAAGAUAAAGUUAAAUACUUC---ACUUACCGCAUUAAUGCCCGAAAGUUGCUGACCACCAUGGAAACAGCAGACAAUGAUCAGGGGCAAUG ---------..(((..(((...((((.....)))).---.)))))).......(((((((..(((((((.......((....)).))).))))..))..)))))... ( -20.70, z-score = -0.52, R) >droEre2.scaffold_4929 7986295 95 + 26641161 ---------AAGGCCCAAGAAAAAGGUGAUUACUCC---ACUUACCGCAUUAAUGCCCGACAGUUGCUGACCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAAUG ---------...((((..((....(((((.......---..)))))(((....)))......((((((..((.....)).)))))).........))..)))).... ( -24.60, z-score = -0.98, R) >droAna3.scaffold_13266 16289553 95 - 19884421 ------------AUUUCAUUUAAAAACAUAAAACUCUAUACCAACCGCAUUGAUGCCGGACAGUUGCUGGCCUCCAUGGAAGCAACAGACAAUGAUCAAGGGAAGAG ------------.....................((((...((..(((((....)).)))...((((((..((.....)).)))))).............))..)))) ( -20.60, z-score = -1.33, R) >dp4.chr3 11950122 105 + 19779522 AAAAUCCGAUAUAAAUAUGUUUUAAGCGAAUAUCAC--UACUCACUGCAUUGAUGCCCGACAACUGCUGGCCGCCGUGGAAGCAGCAGACAAUGACAAGCGGUAGCA .......(((((.....(((.....))).))))).(--(((.(.((.(((((.((.....)).((((((.((.....))...)))))).)))))...)).))))).. ( -20.70, z-score = 0.89, R) >droPer1.super_4 5422692 105 + 7162766 AAAAUCCGAUAUAAAUAUGUUUUAAACGAAUAUCAC--UACUCACUGCAUUGAUGCCCGACAACUGCUGGCCGCCGUGGAAGCAGCAGACAAUGACAAGCGGUAGCA .......(((((.................))))).(--(((.(.((.(((((.((.....)).((((((.((.....))...)))))).)))))...)).))))).. ( -20.63, z-score = 0.43, R) >droWil1.scaffold_181141 3002246 102 + 5303230 ---CUAAUUAAAAAUCAACUUAUUGGUAUCCACAAC--AACUUACCGCAUUAAUUCCGGACAAUUGUUGACCGCCAUGGAAGCAGCAGACAAUGAAUAUGGGCAAUA ---.................(((((.......((((--((....(((.(.....).)))....))))))....(((((....((........))..))))).))))) ( -14.00, z-score = 1.05, R) >droVir3.scaffold_12875 14869866 88 - 20611582 -----------------CAAAACAAAUCAAUUAGAA--AACUCACCGCAUUGAUGCCCGACAGCUGCUGGCCACCCUGGAAGCAGCAGACAAUGAAGAGCGGCAUCA -----------------...................--............((((((((....((((((..((.....)).))))))...(......).).))))))) ( -21.90, z-score = -1.31, R) >droMoj3.scaffold_6496 19842946 93 + 26866924 ----------GAGACCAAAUUGAU-UCAUAAGCUGC---ACUUACUGCAUUGAUGCCCGACAGCUGCUGGCCGCCCUGGAAGCAGCAGACAAUGAUGAGCGGCAGGA ----------..............-.......((((---.((((...(((((.((.....))((((((..((.....)).))))))...))))).))))..)))).. ( -23.60, z-score = 0.63, R) >droGri2.scaffold_15245 14891184 74 - 18325388 -------------------------------AUCAA--UACUUACCGCAUUGAUGCCCGACAAUUGUUGGCCGCCCUGGAAGCAGCAGACAAUGAUGAGCGGCAUCA -------------------------------.....--............((((((((....((((((.((.((.......)).)).)))))).....).))))))) ( -20.20, z-score = -0.84, R) >anoGam1.chr2L 12132608 107 - 48795086 ACGAACAGAACUAUUAAUGAUGGCACUCGCGUCCUCUUGCCCUACCGCAUUGAUUCCGGACAGCUGUUGGCCACCUUGUAAGGCACAGACCAGCACCAGUGGAAGCA ......................((..((((((((((.(((......)))..))....)))).((((((((((.........))).)))..))))....))))..)). ( -23.30, z-score = 1.39, R) >consensus _________AAGGAUCAAGAUAAAGGUGAAUACUAC___ACUUACCGCAUUGAUGCCCGACAGCUGCUGGCCACCAUGGAAGCAGCAGACAAUGAUCAGGGGCAACA .....................................................(((((..((.((((((.((.....))...))))))....))....).))))... (-10.55 = -10.61 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:11 2011