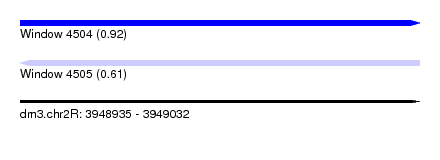
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,948,935 – 3,949,032 |
| Length | 97 |
| Max. P | 0.916430 |

| Location | 3,948,935 – 3,949,032 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Shannon entropy | 0.20846 |
| G+C content | 0.37216 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
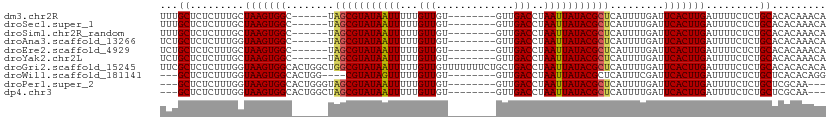
>dm3.chr2R 3948935 97 + 21146708 UUUGCUCUCUUUGCUAAGUGGC------UAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ..(((.........(((((((.------..((((((((((...(((..--------...)))..))))))))))((.....)).))))))).........)))........ ( -19.07, z-score = -1.72, R) >droSec1.super_1 1592586 97 + 14215200 UUUGCUCUCUUUGCUAAGUGGC------UAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ..(((.........(((((((.------..((((((((((...(((..--------...)))..))))))))))((.....)).))))))).........)))........ ( -19.07, z-score = -1.72, R) >droSim1.chr2R_random 1123718 97 + 2996586 UUUGCUCUCUUUGCUAAGUGGC------UAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ..(((.........(((((((.------..((((((((((...(((..--------...)))..))))))))))((.....)).))))))).........)))........ ( -19.07, z-score = -1.72, R) >droAna3.scaffold_13266 16267758 97 + 19884421 UCUGCUCUCUUUGGUAAGUGGC------UAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ..(((.........(((((((.------..((((((((((...(((..--------...)))..))))))))))((.....)).))))))).........)))........ ( -20.07, z-score = -1.78, R) >droEre2.scaffold_4929 7960847 97 - 26641161 UCUGCUCUCUUUGCUAAGUGGC------UAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ..(((.........(((((((.------..((((((((((...(((..--------...)))..))))))))))((.....)).))))))).........)))........ ( -18.97, z-score = -1.74, R) >droYak2.chr2L 16605155 97 + 22324452 UCUGCUCUCUUUGCUAAGUGGC------UAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ..(((.........(((((((.------..((((((((((...(((..--------...)))..))))))))))((.....)).))))))).........)))........ ( -18.97, z-score = -1.74, R) >droGri2.scaffold_15245 14866175 111 + 18325388 UUCGCUCUCUUUGGUAAGUGGCACUGGCUGGCGUAUAAUUUUUGUUGUUUUUUUCUGCUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACACACA ...((.........((((((((....)).(((((((((((...(((((........)).)))..)))))))))))..........)))))).........))......... ( -21.87, z-score = -1.52, R) >droWil1.scaffold_181141 2975569 96 - 5303230 ---GCUCUCUUUGGUAAGUGGCACUGG----CGUAUAGUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUCGAUUCACUUGAUUUUCUCUGCUCACACAGG ---..............((((((..((----(((((((((...(((..--------...)))..)))))))))))....((((.....)))).......))).)))..... ( -19.60, z-score = -0.63, R) >droPer1.super_2 5771064 97 + 9036312 ---GCUCUCUUUGGUAAGUGGCACUGGGUAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCUCGCAA--- ---((.........(((((((....(((((((((((((((...(((..--------...)))..))))))))))).))))....))))))).........))......--- ( -22.27, z-score = -1.70, R) >dp4.chr3 5576396 97 + 19779522 ---GCUCUCUUUGGUAAGUGGCACUGGCUAGCGUAUAAUUUUUGUUGU--------GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCUCGCAA--- ---((.........((((((((....)).(((((((((((...(((..--------...)))..)))))))))))..........)))))).........))......--- ( -20.87, z-score = -1.38, R) >consensus UUUGCUCUCUUUGCUAAGUGGC______UAGCGUAUAAUUUUUGUUGU________GUUGACCUAAUUAUACGCUCAUUUUGAUUCACUUGAUUUUCUCUGCACACAAACA ...((.........(((((((........(((((((((((...((((...........))))..))))))))))).........))))))).........))......... (-18.65 = -18.68 + 0.03)
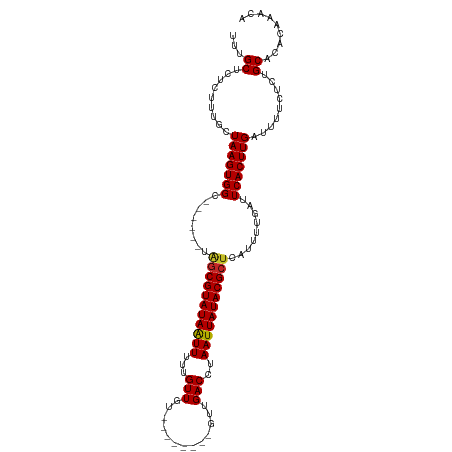
| Location | 3,948,935 – 3,949,032 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Shannon entropy | 0.20846 |
| G+C content | 0.37216 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3948935 97 - 21146708 UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUA------GCCACUUAGCAAAGAGAGCAAA (((((...(((.((.....))(((((..((.....))((((((((((..((....--------...))...))))))))))..------..))))).)))....))))).. ( -21.90, z-score = -2.59, R) >droSec1.super_1 1592586 97 - 14215200 UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUA------GCCACUUAGCAAAGAGAGCAAA (((((...(((.((.....))(((((..((.....))((((((((((..((....--------...))...))))))))))..------..))))).)))....))))).. ( -21.90, z-score = -2.59, R) >droSim1.chr2R_random 1123718 97 - 2996586 UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUA------GCCACUUAGCAAAGAGAGCAAA (((((...(((.((.....))(((((..((.....))((((((((((..((....--------...))...))))))))))..------..))))).)))....))))).. ( -21.90, z-score = -2.59, R) >droAna3.scaffold_13266 16267758 97 - 19884421 UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUA------GCCACUUACCAAAGAGAGCAGA .((..(((.((.((.....))...............(((((((((((..((....--------...))...))))))))))).------)))))..))............. ( -18.90, z-score = -1.58, R) >droEre2.scaffold_4929 7960847 97 + 26641161 UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUA------GCCACUUAGCAAAGAGAGCAGA (((((...(((.((.....))(((((..((.....))((((((((((..((....--------...))...))))))))))..------..))))).)))....))))).. ( -21.90, z-score = -2.40, R) >droYak2.chr2L 16605155 97 - 22324452 UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUA------GCCACUUAGCAAAGAGAGCAGA (((((...(((.((.....))(((((..((.....))((((((((((..((....--------...))...))))))))))..------..))))).)))....))))).. ( -21.90, z-score = -2.40, R) >droGri2.scaffold_15245 14866175 111 - 18325388 UGUGUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAGCAGAAAAAAACAACAAAAAUUAUACGCCAGCCAGUGCCACUUACCAAAGAGAGCGAA .(((.(((.(((((.....)).............((.((((((((((..((...............))...)))))))))))).....)))))).)))............. ( -18.26, z-score = -0.41, R) >droWil1.scaffold_181141 2975569 96 + 5303230 CCUGUGUGAGCAGAGAAAAUCAAGUGAAUCGAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAACUAUACG----CCAGUGCCACUUACCAAAGAGAGC--- .((((....))))........(((((...((...((.(((((((.((..((....--------...))...)).))))))----))).)).)))))............--- ( -15.00, z-score = -0.13, R) >droPer1.super_2 5771064 97 - 9036312 ---UUGCGAGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUACCCAGUGCCACUUACCAAAGAGAGC--- ---......((.((.....))(((((...((.....(((((((((((..((....--------...))...)))))))))))......)).)))))..........))--- ( -16.90, z-score = -1.70, R) >dp4.chr3 5576396 97 - 19779522 ---UUGCGAGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC--------ACAACAAAAAUUAUACGCUAGCCAGUGCCACUUACCAAAGAGAGC--- ---......((.((.....))(((((...((.....(((((((((((..((....--------...))...)))))))))))......)).)))))..........))--- ( -16.90, z-score = -1.24, R) >consensus UGUUUGUGUGCAGAGAAAAUCAAGUGAAUCAAAAUGAGCGUAUAAUUAGGUCAAC________ACAACAAAAAUUAUACGCUA______GCCACUUACCAAAGAGAGCAAA .........((.((.....))(((((..........(((((((((((..((...............))...))))))))))).........)))))..........))... (-14.90 = -15.30 + 0.40)

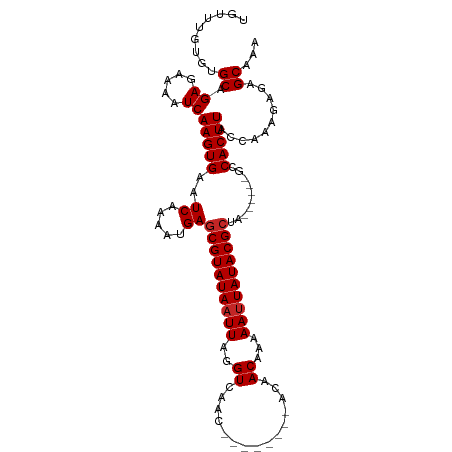
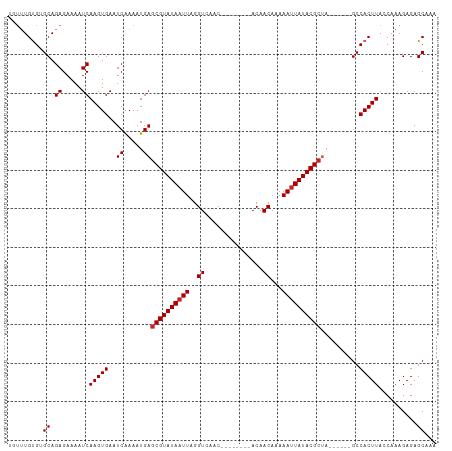
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:07 2011