
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,928,727 – 3,928,833 |
| Length | 106 |
| Max. P | 0.520310 |

| Location | 3,928,727 – 3,928,833 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Shannon entropy | 0.48686 |
| G+C content | 0.70665 |
| Mean single sequence MFE | -50.99 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
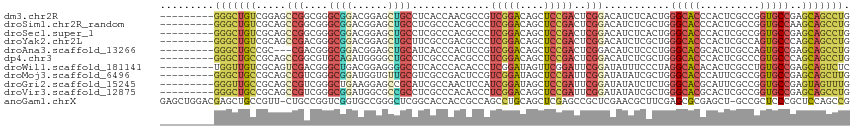
>dm3.chr2R 3928727 106 + 21146708 ---------GGGCUGUCGGAGCCGGCGGGCGGACGGAGCUGCCUCACCAACGCCGUCGGACAGCUCCGACUCGGACAUCUCACUGGGCACCCACUCGCCGGUGCCGAGCAGCCUG ---------((((((((((.(((((((((.((.((((((((((..............)).)))))))).(((((........)))))...)).))))))))).)))).)))))). ( -60.34, z-score = -3.72, R) >droSim1.chr2R_random 1116718 106 + 2996586 ---------GGGCUGUCGCAGCCGGCGGGCGGACGGAGCUGCCUCGCCCACGCCCUCGGACAGCUCCGACUCGGACAUCUCGCUGGGCACCCACUCGCCGGUGCCAAGCAGCCUG ---------(((((((.(((.((((((((.((.((((((((((..((....))....)).)))))))).(((((........)))))...)).)))))))))))...))))))). ( -56.80, z-score = -2.45, R) >droSec1.super_1 1567836 106 + 14215200 ---------GGGCUGUCGCAGCCGGCGGGCGGACGGAGCUGCCUCGCCCACGCCCUCGGACAGCUCCGACUCGGACAUCUCACUGGGCACCCACUCGCCGGUGCCGAGCAGCCUG ---------(((((((((..(((((((((.((.((((((((((..((....))....)).)))))))).(((((........)))))...)).)))))))))..))).)))))). ( -57.50, z-score = -2.70, R) >droYak2.chr2L 16584528 106 + 22324452 ---------GGGCUGUCGCAGCCGACGGGCGGACGGAGCUGCUUCGCCGACGCCCUCGGACAGCUCCGACUCGGACAUCUCGCUGGGCACCCACUCGCCAGUGCCCAGCAGCCUG ---------((((((((...(((....)))...((((((((.....((((.....)))).)))))))).....))).....(((((((((..........)))))))))))))). ( -52.60, z-score = -1.98, R) >droAna3.scaffold_13266 16248277 103 + 19884421 ---------GGGCUGCCGC---CGACGGGCGGACGGAGCUGCAUCACCCACUCCGUCGGACAGCUCCGACUCGGACAUCUCCCUGGGCACGCACUCGCCAGUGCCGAGCAGCCUG ---------((((((((((---(....))))).(((((((((...((.......))..).)))))))).((((((....)))...(((((((....))..)))))))))))))). ( -46.20, z-score = -1.25, R) >dp4.chr3 5557648 106 + 19779522 ---------GGGCUGCCGCAGCCGGCGUGCAGAUGGGGCUGCCUCGCCCACGCCCUCGGACAGCUCCGACUCGGACAUCUCGCUGGGCACCCACUCGCCCGUGCCCAGCAGCCUG ---------(((((((((....))))..(.(((((((((......)))).((...(((((....)))))..))..))))))(((((((((..........)))))))))))))). ( -50.00, z-score = -0.89, R) >droWil1.scaffold_181141 2361528 106 + 5303230 ---------UGGUUGUCGCAGUCGACGGGCUGACGGAGGGGCCUCACCCACACCCUCGGAUAGUUCGGAUUCGGAUAUUUCCCUAGGCACACACUCGCCUGUGCCGAGCAGUCUC ---------.((.((((((((((....))))).))..(((......)))))).))..((((.((((((....(((....))).(((((........)))))..)))))).)))). ( -36.90, z-score = 0.13, R) >droMoj3.scaffold_6496 19787636 106 - 26866924 ---------GGGCUGCCGCAGCCGUCGGGCGGAUGGUGUUGCGUCGCCGACUCCGUCGGAUAGCUCCGAUUCGGAUAUAUCGCUGGGCACCCAUUCGCCGGUGCCGAGCAGCUUG ---------((((((((((.((..((.(((((((((((......)))))..)))))).))..))((((...))))......))..((((((........))))))).))))))). ( -49.00, z-score = -0.94, R) >droGri2.scaffold_15245 14841542 106 + 18325388 ---------GGGUUGCCGCAGCCGUCGGGCUGAAGGAGCCGCAUCGCCAACUCCAUCGGAUAGCUCCGAUUCGGAUAUAUCUCUGGGCACGCAUUCGCCGGUGCCGAGUAGUUUG ---------.((((.(..(((((....)))))..).))))((((((((......((((((....)))))).((((......)))))))..((....)).)))))........... ( -36.80, z-score = 0.51, R) >droVir3.scaffold_12875 14826902 106 + 20611582 ---------GGGCUGCCGCAGCCGUCGGGCGGAUGGCGCCGCCUCGCCCACACCCUCGGACAGCUCCGAUUCGGAUAUAUCGCUGGGCACGCACUCGCCGGUGCCGAGCAGCCUG ---------((((((((.((((.((.((((((..(((...))))))))))).((.(((((....)))))...)).......))))(((((((....))..)))))).))))))). ( -49.00, z-score = -0.58, R) >anoGam1.chrX 9832119 113 + 22145176 GAGCUGGACGAGCUGCCGUU-CUGCCGGUCGGUGCCGGGCUCGGCACCACCGCCAGCCUGCAGCUCGAGCCGCUCGAACGCUUCGAGCGCGAGCU-GCCGCUCCCGCUCCAGCCG (.((((((((((((((.(..-(((.((((.(((((((....))))))))))).))).).)))))))).(((((((((.....))))))).((((.-...))))..))))))))). ( -65.80, z-score = -3.55, R) >consensus _________GGGCUGCCGCAGCCGGCGGGCGGACGGAGCUGCCUCGCCCACGCCCUCGGACAGCUCCGACUCGGACAUCUCGCUGGGCACCCACUCGCCGGUGCCGAGCAGCCUG .........(((((((.....(((....((((......)))).............(((((....)))))..)))...........(((((..........)))))..))))))). (-22.85 = -23.32 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:04 2011