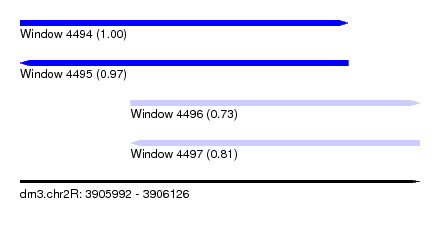
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,905,992 – 3,906,126 |
| Length | 134 |
| Max. P | 0.999228 |

| Location | 3,905,992 – 3,906,102 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.62997 |
| G+C content | 0.47316 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -12.76 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
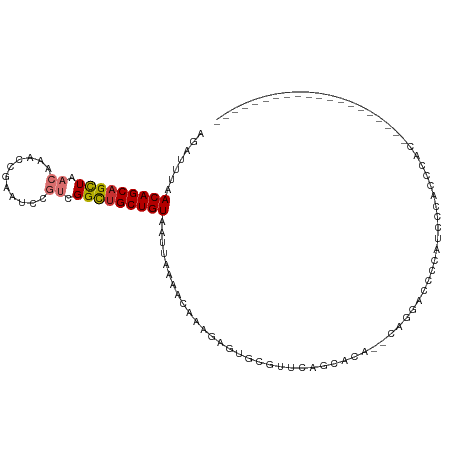
>dm3.chr2R 3905992 110 + 21146708 AGAUUUAACAGCAGCUAACAAACCGAAUUCGUCGGCUGCUGUAAUUAAAACAAAGAGUGCGUUCAGCACA--CAGGACCCCAUCCCACGCACUUCCUUCCUCCCCUCCUCUU .......((((((((..((...........))..))))))))............((((((((........--..(((.....))).)))))))).................. ( -25.60, z-score = -3.32, R) >droPer1.super_2 5732082 88 + 9036312 AGACCUAACAGCAGCUAACCAAACCAAGAAACUGGCUGCUGUAAUUAAA-CAGAUGCUAAGAGUAGCACA--CAGACUCCCGCCUCAGCCC--------------------- .......((((((((((...............)))))))))).......-..((.((...((((......--...))))..)).)).....--------------------- ( -19.76, z-score = -1.82, R) >dp4.chr3 5537866 88 + 19779522 AGACCUAACAGCAGCUAACCAAACCAAGAAACUGGCUGCUGUAAUUAAA-CAGAUGCUAAGAGUAGCACA--CAGACUCCCGCCUCAGCCC--------------------- .......((((((((((...............)))))))))).......-..((.((...((((......--...))))..)).)).....--------------------- ( -19.76, z-score = -1.82, R) >droAna3.scaffold_13266 16223715 90 + 19884421 GGAUUUAACAGCAGUUAACAAACCAAGCCCGUCGGUGGCUGUAAUUAAAACAAAAAAAACAGAGAUAGCAGCCUUAUCUUCUUUCUCUAU---------------------- .......(((((.(((....)))...(((....))).)))))..................(((((.((.((......)).)).)))))..---------------------- ( -14.20, z-score = -0.30, R) >droSim1.chr2R_random 1107514 110 + 2996586 AGAUUUAACAGCAGCUAACAAACCGAAUCCGUCGGCUGCUGUAAUUAAAACAAAGAGUGCGUUCAGCACA--CAGGACCCCAUCCCUCGCACUUCCUUCCUCCCCUCCUCUU .......((((((((..((...........))..))))))))............(((((((.........--.(((........)))))))))).................. ( -25.40, z-score = -3.18, R) >droSec1.super_1 1542138 110 + 14215200 AGAUUUAACAGCAGCUAACAAACCGAAUCCGUCGGCUGCUGUAAUUAAAACAAAGAGUGCGUUCAGCACA--CAGGACCCCAUCCCUCGCACUUCCUUCCUCCCCUCCUCUU .......((((((((..((...........))..))))))))............(((((((.........--.(((........)))))))))).................. ( -25.40, z-score = -3.18, R) >droYak2.chr2L 16561376 90 + 22324452 AGAUUUAACAGCAGCUAACAAACCGAAUCCGUCGGCUGCUGUAAUUAAAACAAAGAGUGCGUUCAGCACA--CAGGACCUCAUCCUUCUCAA-------------------- .......((((((((..((...........))..))))))))...........(((((((.....)))).--.((((.....)))))))...-------------------- ( -24.80, z-score = -3.45, R) >droEre2.scaffold_4929 7919336 97 - 26641161 AGAUUUAACAGCAGCUAACAAACCGAAUCCGUCGGCUGCUGUAAUUAAAACAAAGAGUGCGUUCCGCACAUCCUGUGUGCAUCCCCAGCUUCCUCGG--------------- .......((((((((..((...........))..))))))))............(((.((.....((((((...)))))).......))...)))..--------------- ( -26.30, z-score = -2.60, R) >consensus AGAUUUAACAGCAGCUAACAAACCGAAUCCGUCGGCUGCUGUAAUUAAAACAAAGAGUGCGUUCAGCACA__CAGGACCCCAUCCCACCCAC____________________ .......(((((((((.((...........)).)))))))))...................................................................... (-12.76 = -13.16 + 0.41)

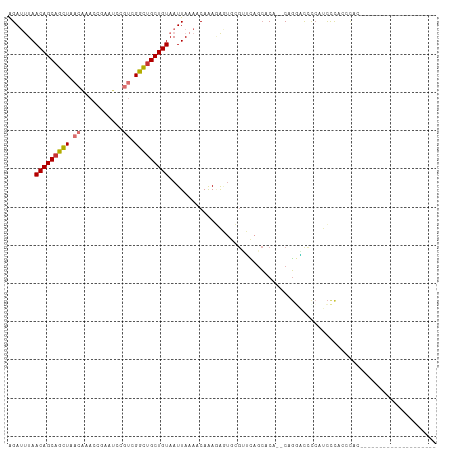
| Location | 3,905,992 – 3,906,102 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.62997 |
| G+C content | 0.47316 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.19 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3905992 110 - 21146708 AAGAGGAGGGGAGGAAGGAAGUGCGUGGGAUGGGGUCCUG--UGUGCUGAACGCACUCUUUGUUUUAAUUACAGCAGCCGACGAAUUCGGUUUGUUAGCUGCUGUUAAAUCU ..((((..(((((...(..((..((..((((...))))..--))..))...)...)))))..))))....((((((((..((((((...))))))..))))))))....... ( -40.40, z-score = -3.24, R) >droPer1.super_2 5732082 88 - 9036312 ---------------------GGGCUGAGGCGGGAGUCUG--UGUGCUACUCUUAGCAUCUG-UUUAAUUACAGCAGCCAGUUUCUUGGUUUGGUUAGCUGCUGUUAGGUCU ---------------------(((((.((((....)))).--.((((((....))))))...-..((((..((((((((((.........)))))).))))..))))))))) ( -25.50, z-score = 0.24, R) >dp4.chr3 5537866 88 - 19779522 ---------------------GGGCUGAGGCGGGAGUCUG--UGUGCUACUCUUAGCAUCUG-UUUAAUUACAGCAGCCAGUUUCUUGGUUUGGUUAGCUGCUGUUAGGUCU ---------------------(((((.((((....)))).--.((((((....))))))...-..((((..((((((((((.........)))))).))))..))))))))) ( -25.50, z-score = 0.24, R) >droAna3.scaffold_13266 16223715 90 - 19884421 ----------------------AUAGAGAAAGAAGAUAAGGCUGCUAUCUCUGUUUUUUUUGUUUUAAUUACAGCCACCGACGGGCUUGGUUUGUUAACUGCUGUUAAAUCC ----------------------(((((((.((.((......)).)).)))))))................(((((....(((((((...)))))))....)))))....... ( -17.60, z-score = 0.05, R) >droSim1.chr2R_random 1107514 110 - 2996586 AAGAGGAGGGGAGGAAGGAAGUGCGAGGGAUGGGGUCCUG--UGUGCUGAACGCACUCUUUGUUUUAAUUACAGCAGCCGACGGAUUCGGUUUGUUAGCUGCUGUUAAAUCU ..((((..(((((...(..((..((.(((((...))))).--))..))...)...)))))..))))....((((((((..((((((...))))))..))))))))....... ( -37.30, z-score = -2.11, R) >droSec1.super_1 1542138 110 - 14215200 AAGAGGAGGGGAGGAAGGAAGUGCGAGGGAUGGGGUCCUG--UGUGCUGAACGCACUCUUUGUUUUAAUUACAGCAGCCGACGGAUUCGGUUUGUUAGCUGCUGUUAAAUCU ..((((..(((((...(..((..((.(((((...))))).--))..))...)...)))))..))))....((((((((..((((((...))))))..))))))))....... ( -37.30, z-score = -2.11, R) >droYak2.chr2L 16561376 90 - 22324452 --------------------UUGAGAAGGAUGAGGUCCUG--UGUGCUGAACGCACUCUUUGUUUUAAUUACAGCAGCCGACGGAUUCGGUUUGUUAGCUGCUGUUAAAUCU --------------------...(((((((..(((....(--((((.....))))).)))..))))....((((((((..((((((...))))))..))))))))....))) ( -29.00, z-score = -2.37, R) >droEre2.scaffold_4929 7919336 97 + 26641161 ---------------CCGAGGAAGCUGGGGAUGCACACAGGAUGUGCGGAACGCACUCUUUGUUUUAAUUACAGCAGCCGACGGAUUCGGUUUGUUAGCUGCUGUUAAAUCU ---------------((.((....)).))(((..(((.((((.(((((...)))))))))))).......((((((((..((((((...))))))..))))))))...))). ( -33.40, z-score = -2.46, R) >consensus ____________________GUGCGUGGGAUGGGGUCCUG__UGUGCUGAACGCACUCUUUGUUUUAAUUACAGCAGCCGACGGAUUCGGUUUGUUAGCUGCUGUUAAAUCU ...........................((((...........((((.....))))...............((((((((.((((((.....)))))).))))))))...)))) (-15.62 = -15.19 + -0.43)
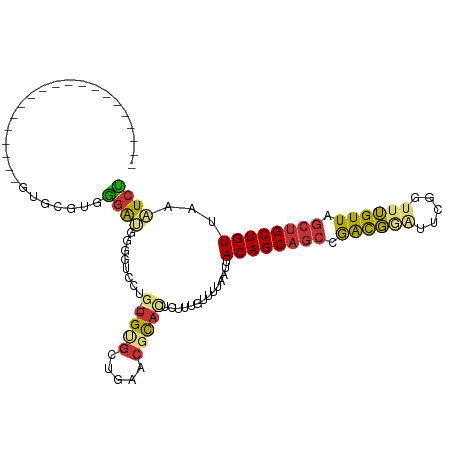
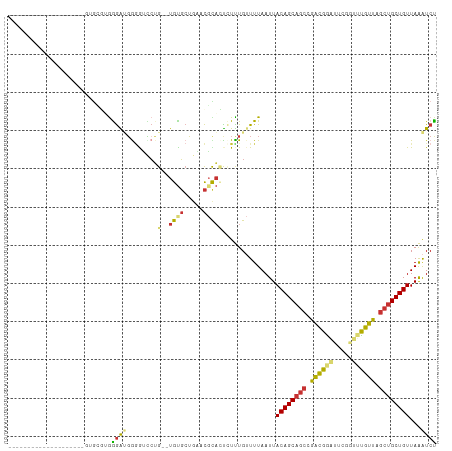
| Location | 3,906,029 – 3,906,126 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 61.92 |
| Shannon entropy | 0.59441 |
| G+C content | 0.47289 |
| Mean single sequence MFE | -13.57 |
| Consensus MFE | -4.82 |
| Energy contribution | -4.26 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
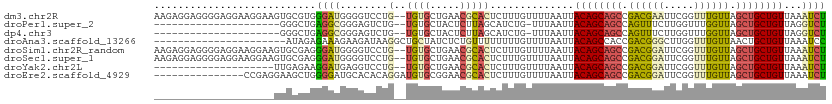
>dm3.chr2R 3906029 97 + 21146708 GCUGUAAUUAAAACAAAGAGUGCGUUCAGCACACAGGACCCCAUCCCACGCACUUCCUUCCUCCCCUCCUCUUUGUCCCCACAAAAGCAACGCAUAU----- (((.........(((((((((((.....))))...(((.....))).......................))))))).........))).........----- ( -17.57, z-score = -3.26, R) >droAna3.scaffold_13266 16223752 79 + 19884421 GCUGUAAUUAAAACAAAAAAAACAGAGAUAGCA----GCCUUAUCUUCUUUCUCUAUUUUCUUCAAAGAACCGUAGCAC-AUGU------------------ ((((...................(((((.((.(----(......)).)).)))))...((((....))))...))))..-....------------------ ( -10.40, z-score = -0.83, R) >droSim1.chr2R_random 1107551 102 + 2996586 GCUGUAAUUAAAACAAAGAGUGCGUUCAGCACACAGGACCCCAUCCCUCGCACUUCCUUCCUCCCCUCCUCUUCGUCCCCACGUAGUCCGUGCAACGCAUAU ...................(((((((..((((...(((.....)))........................((.(((....))).))...))))))))))).. ( -16.70, z-score = -1.86, R) >droSec1.super_1 1542175 83 + 14215200 GCUGUAAUUAAAACAAAGAGUGCGUUCAGCACACAGGACCCCAUCCCUCGCACUUCCUUCCUCCCCUCCUCUUCGUCCC-AUAU------------------ ............((.((((((((.....))))...(((.....))).......................)))).))...-....------------------ ( -9.60, z-score = -1.15, R) >consensus GCUGUAAUUAAAACAAAGAGUGCGUUCAGCACACAGGACCCCAUCCCUCGCACUUCCUUCCUCCCCUCCUCUUCGUCCC_ACAU__________________ .................(((((((........................)))))))............................................... ( -4.82 = -4.26 + -0.56)

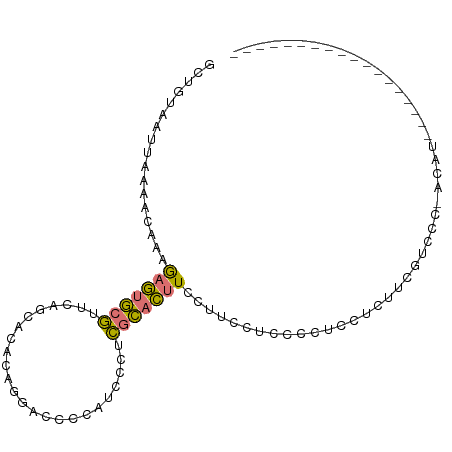
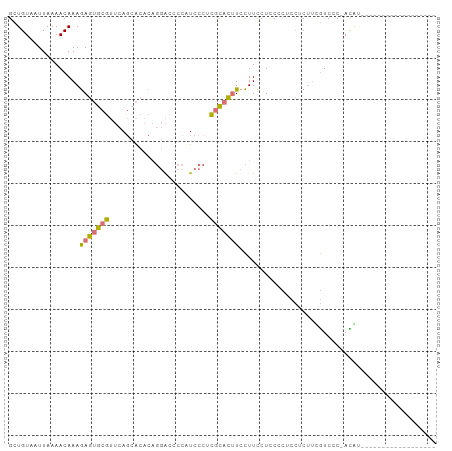
| Location | 3,906,029 – 3,906,126 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 61.92 |
| Shannon entropy | 0.59441 |
| G+C content | 0.47289 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -10.71 |
| Energy contribution | -11.53 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3906029 97 - 21146708 -----AUAUGCGUUGCUUUUGUGGGGACAAAGAGGAGGGGAGGAAGGAAGUGCGUGGGAUGGGGUCCUGUGUGCUGAACGCACUCUUUGUUUUAAUUACAGC -----......(((((((((.((....)).))))).((((((...(..((..((..((((...))))..))..))...)...))))))..........)))) ( -27.80, z-score = -1.51, R) >droAna3.scaffold_13266 16223752 79 - 19884421 ------------------ACAU-GUGCUACGGUUCUUUGAAGAAAAUAGAGAAAGAAGAUAAGGC----UGCUAUCUCUGUUUUUUUUGUUUUAAUUACAGC ------------------...(-((((....)).....((((((((((((((.((.((......)----).)).)))))))))))))).........))).. ( -17.60, z-score = -1.28, R) >droSim1.chr2R_random 1107551 102 - 2996586 AUAUGCGUUGCACGGACUACGUGGGGACGAAGAGGAGGGGAGGAAGGAAGUGCGAGGGAUGGGGUCCUGUGUGCUGAACGCACUCUUUGUUUUAAUUACAGC ......((((((((.....))))(((((((((((..(.(.........((..((.(((((...))))).))..))...).).))))))))))).....)))) ( -28.50, z-score = -1.04, R) >droSec1.super_1 1542175 83 - 14215200 ------------------AUAU-GGGACGAAGAGGAGGGGAGGAAGGAAGUGCGAGGGAUGGGGUCCUGUGUGCUGAACGCACUCUUUGUUUUAAUUACAGC ------------------...(-(((((((((((..(.(.........((..((.(((((...))))).))..))...).).))))))))))))........ ( -22.70, z-score = -1.99, R) >consensus __________________ACAU_GGGACAAAGAGGAGGGGAGGAAGGAAGUGCGAGGGAUGGGGUCCUGUGUGCUGAACGCACUCUUUGUUUUAAUUACAGC .......................(((((((((((........(.....((((((..(((.....)))..)))))).....).)))))))))))......... (-10.71 = -11.53 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:07:00 2011