| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,875,040 – 3,875,144 |
| Length | 104 |
| Max. P | 0.959130 |

| Location | 3,875,040 – 3,875,144 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.44 |
| Shannon entropy | 0.25462 |
| G+C content | 0.41477 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.949908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
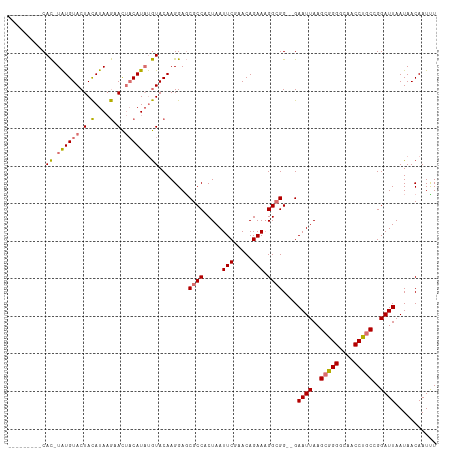
>dm3.chr2R 3875040 104 + 21146708 ---------CAC-UAUGUACUACAUAAGAACUACAUAUGUACAAGGAGCGCCACUAAUUCGAACAGAAAGGCGGUCGAAUUAAGCGGGGCAACCUGCCGGAUUAAUAACAAUUU ---------..(-(.(((((..................)))))))((.((((.....(((.....))).)))).)).((((..(((((....)))))..))))........... ( -25.57, z-score = -2.63, R) >droEre2.scaffold_4929 7887738 112 - 26641161 UGUUCACCACAUGUGUGUACUAUAUACAAAU--CAUCCAUACAAGGAUCGCCCCUAAUUCAAACAGAAUGGCGGUCGAAUUAAGCAGGGAAACCUACGGGAUUAAUAACAAUAU ((((.......(((((((...)))))))...--.((((.......(((((((....((((.....))))))))))).......(.(((....))).).))))....)))).... ( -26.30, z-score = -1.80, R) >droSec1.super_1 1512296 102 + 14215200 ---------CAC-UAUGUACUACAUAAGUACUACAUAUGUACAAGGAGCGCCACUAAUUCGAACAGAAAGGAGG--GAAUUAAGCGGGGCAACCUGCCGGAUUAAUAACAAUUU ---------.((-((((((.(((....))).)))))).))..........((.((..(((.....)))...)))--)((((..(((((....)))))..))))........... ( -22.70, z-score = -1.96, R) >droSim1.chr2R 2582228 102 + 19596830 ---------CAC-UAUGUACUACAUAAGUACUACAUAUGUACAAGGAGCGCCACUAAUUCGAACAGAAAGGCGG--GAAUUAAGCGGGGCAACCUGCGGGAUUAAUAACAAUCU ---------.((-((((((.(((....))).)))))).))....(((.((((.....(((.....))).)))).--.((((..(((((....)))))..))))........))) ( -26.20, z-score = -2.89, R) >consensus _________CAC_UAUGUACUACAUAAGAACUACAUAUGUACAAGGAGCGCCACUAAUUCGAACAGAAAGGCGG__GAAUUAAGCGGGGCAACCUGCCGGAUUAAUAACAAUUU ..........((.((((((.(((....))).)))))).))........((((.....(((.....))).))))....((((..(((((....)))))..))))........... (-19.10 = -19.73 + 0.62)


| Location | 3,875,040 – 3,875,144 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Shannon entropy | 0.25462 |
| G+C content | 0.41477 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.75 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
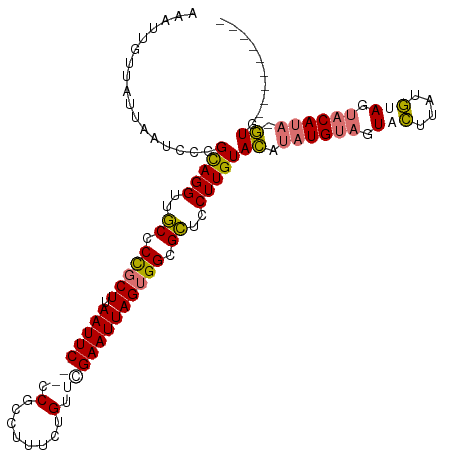
>dm3.chr2R 3875040 104 - 21146708 AAAUUGUUAUUAAUCCGGCAGGUUGCCCCGCUUAAUUCGACCGCCUUUCUGUUCGAAUUAGUGGCGCUCCUUGUACAUAUGUAGUUCUUAUGUAGUACAUA-GUG--------- .................(((((..((.(((((.(((((((.((......)).)))))))))))).))..)))))((.((((((.(.(....).).))))))-)).--------- ( -27.30, z-score = -2.62, R) >droEre2.scaffold_4929 7887738 112 + 26641161 AUAUUGUUAUUAAUCCCGUAGGUUUCCCUGCUUAAUUCGACCGCCAUUCUGUUUGAAUUAGGGGCGAUCCUUGUAUGGAUG--AUUUGUAUAUAGUACACACAUGUGGUGAACA .((((((...((((((((((((....))))).((((((((.((......)).))))))))))))..((((......)))).--..)))...))))))(((....)))....... ( -24.30, z-score = 0.27, R) >droSec1.super_1 1512296 102 - 14215200 AAAUUGUUAUUAAUCCGGCAGGUUGCCCCGCUUAAUUC--CCUCCUUUCUGUUCGAAUUAGUGGCGCUCCUUGUACAUAUGUAGUACUUAUGUAGUACAUA-GUG--------- .................(((((..((.(((((.(((((--...(......)...)))))))))).))..)))))((.((((((.(((....))).))))))-)).--------- ( -26.10, z-score = -2.99, R) >droSim1.chr2R 2582228 102 - 19596830 AGAUUGUUAUUAAUCCCGCAGGUUGCCCCGCUUAAUUC--CCGCCUUUCUGUUCGAAUUAGUGGCGCUCCUUGUACAUAUGUAGUACUUAUGUAGUACAUA-GUG--------- .(((((....)))))..(((((..((.(((((.(((((--..((......))..)))))))))).))..)))))((.((((((.(((....))).))))))-)).--------- ( -29.50, z-score = -4.04, R) >consensus AAAUUGUUAUUAAUCCCGCAGGUUGCCCCGCUUAAUUC__CCGCCUUUCUGUUCGAAUUAGUGGCGCUCCUUGUACAUAUGUAGUACUUAUGUAGUACAUA_GUG_________ .................(((((..((.(((((.(((((((.(........).)))))))))))).))..)))))((.((((((.(((....))).)))))).)).......... (-20.56 = -21.75 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:57 2011