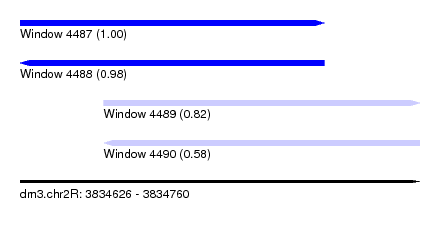
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,834,626 – 3,834,760 |
| Length | 134 |
| Max. P | 0.998772 |

| Location | 3,834,626 – 3,834,728 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Shannon entropy | 0.19806 |
| G+C content | 0.33113 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -26.37 |
| Energy contribution | -27.81 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.02 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.998772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3834626 102 + 21146708 UUAAAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGCCAGGAAGUGAACACAGAGUUCAUUUCAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUAAAA ....((((((((((((((((((.....(--------(.((....)).))...)))))))))))))))))).............((((((((....))))))))....... ( -33.30, z-score = -4.76, R) >droSim1.chr2R 2531008 102 + 19596830 UUGAAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGUCAGGAAAUUAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAAUACAA ...(((((((((((((((((.((((..(--------(.((....)).)).)))))))))))))))))))))............((((((((....))))))))....... ( -29.60, z-score = -3.81, R) >droSec1.super_1 1471585 102 + 14215200 UUGAAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGUCAGGAAAUGAACACAGAGUUCAUUUUAGAUGAAUUCAUGGAAAUUAAAGCUUAUUUCUAUUACAA ...(((((((((((((((((((.(((..--------............))).)))))))))))))))))))..........((((((((........))))))))..... ( -31.34, z-score = -4.18, R) >droYak2.chr2L 16491552 110 + 22324452 UUGAGAAUGAGCUCUGUGUUGAAUUCGCAGAAUUCGCAGAAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAAAUGAAUUCAAGGAAAUUAACGCUUUUUUGUAUUACAA ...(((((((((((((((((.(.(((((.......)).((....))..))).).)))))))))))))))))...(((((.(((((((........))))))).))).)). ( -30.70, z-score = -2.60, R) >droEre2.scaffold_4929 7847440 100 - 26641161 UUGAAAAUGAGCUCUGUGUUUAAUUCGU---------AGAAGUGACAGGA-GUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAACGCUUAUUUCUAUUAUAA ...(((((((((((((((((((.(((..---------..........)))-.)))))))))))))))))))............((((((((....))))))))....... ( -32.10, z-score = -4.77, R) >consensus UUGAAAAUGAGCUCUGUGUUUAAUUCGC________CAGGAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUACAA ...(((((((((((((((((((.(((......................))).)))))))))))))))))))............((((((((....))))))))....... (-26.37 = -27.81 + 1.44)

| Location | 3,834,626 – 3,834,728 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.40 |
| Shannon entropy | 0.19806 |
| G+C content | 0.33113 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3834626 102 - 21146708 UUUUAAUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUGAAAUGAACUCUGUGUUCACUUCCUGGCACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUUUAA ......(((((((.(((.((((((((..((....))...)))))))).((((((((.(.((((..(.....)..--------).))).).))))))))))).))))))). ( -27.60, z-score = -4.43, R) >droSim1.chr2R 2531008 102 - 19596830 UUGUAUUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUAAUUUCCUGACACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUUCAA .....(((....)))(..((((......))))..)....(((((((.((((((((((((((((.(......).)--------).))))).))))))))).)))))))... ( -24.00, z-score = -3.65, R) >droSec1.super_1 1471585 102 - 14215200 UUGUAAUAGAAAUAAGCUUUAAUUUCCAUGAAUUCAUCUAAAAUGAACUCUGUGUUCAUUUCCUGACACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUUCAA ........(((((........))))).............(((((((.(((((((((...(((((.........)--------).)))...))))))))).)))))))... ( -20.80, z-score = -2.47, R) >droYak2.chr2L 16491552 110 - 22324452 UUGUAAUACAAAAAAGCGUUAAUUUCCUUGAAUUCAUUUAAAAUGAACUCUGUGUUCACUUCCUGACACUUCUGCGAAUUCUGCGAAUUCAACACAGAGCUCAUUCUCAA .........................................(((((.(((((((((((.....))).........((((((...)))))).)))))))).)))))..... ( -19.10, z-score = -1.42, R) >droEre2.scaffold_4929 7847440 100 + 26641161 UUAUAAUAGAAAUAAGCGUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUCAC-UCCUGUCACUUCU---------ACGAAUUAAACACAGAGCUCAUUUUCAA ........((((..(((((((((((...((....))....))))))))((((((((...-.....((......---------..))....)))))))))))...)))).. ( -20.80, z-score = -3.63, R) >consensus UUGUAAUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUCACUUCCUGACACUCCUG________GCGAAUUAAACACAGAGCUCAUUUUCAA .......................................(((((((.(((((((((...(((......................)))...))))))))).)))))))... (-16.09 = -16.33 + 0.24)

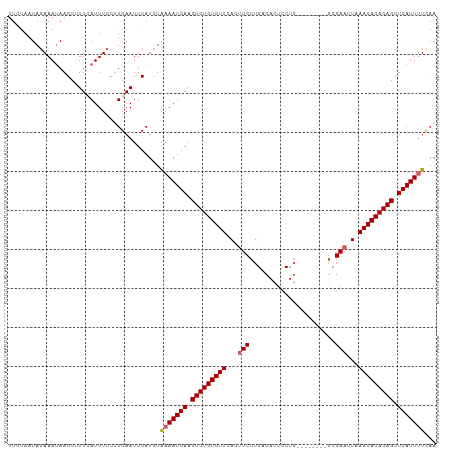
| Location | 3,834,654 – 3,834,760 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Shannon entropy | 0.37906 |
| G+C content | 0.31679 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.02 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3834654 106 + 21146708 CAGGAGUGCCAGGAAGUGAACACAGAGUUCAUUUCAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUAAAAAUUCACCUAUGC-UUGGUAA-GCAUUAUAUGUAA-- ...(..(((((((..(((((....(((((((((...)))))))))..((((((((....))))))))........))))).....)-)))))).-.)...........-- ( -27.50, z-score = -2.69, R) >droSim1.chr2R 2531036 110 + 19596830 CAGGAGUGUCAGGAAAUUAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAAUACAAAUUUACCUAUGCCUUGGUUUUACCUUGCCAUUAUAA ((((.((((.((((((((......(((((((((...)))))))))..((((((((....))))))))......))))).))))))))))).................... ( -23.40, z-score = -1.55, R) >droSec1.super_1 1471613 103 + 14215200 CAGGAGUGUCAGGAAAUGAACACAGAGUUCAUUUUAGAUGAAUUCAUGGAAAUUAAAGCUUAUUUCUAUUACAAAUUCACCUAUGCCUUGGUAA-GCAUUAUAA------ ((((.((((.(((((((((((.....))))))))....((((((.((((((((........))))))))....)))))))))))))))))....-.........------ ( -23.20, z-score = -1.95, R) >droYak2.chr2L 16491588 102 + 22324452 CAGAAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAAAUGAAUUCAAGGAAAUUAACGCUUUUUUGUAUUACAAUUUCACUUAUAC-UUGGUAA-ACAUUAUAA------ ..(((...(((.(((((((((.....)))))))))...))).)))(((((((((((..(......)..)))..))))).)))....-.......-.........------ ( -18.20, z-score = -1.21, R) >droEre2.scaffold_4929 7847467 84 - 26641161 UAGAAGUGACAGGA-GUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAACGCUUAUUUCUAUUAUAAUUUCACCUAUG------------------------- ...........((.-(((((....(((((((((...)))))))))..((((((((....))))))))........)))))))...------------------------- ( -18.30, z-score = -2.13, R) >consensus CAGGAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUACAAAUUCACCUAUGC_UUGGUAA_ACAUUAUAA______ ..(((...(((.(((((((((.....)))))))))...))).)))..((((((((....))))))))........................................... (-13.06 = -14.02 + 0.96)

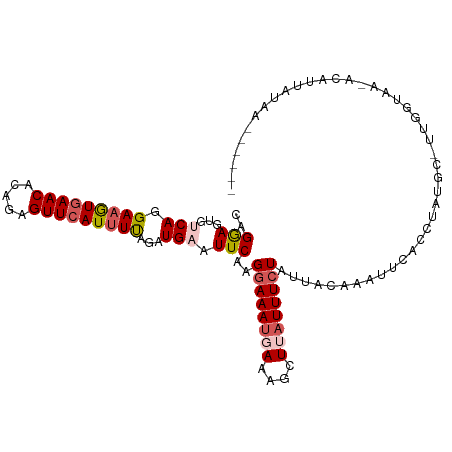
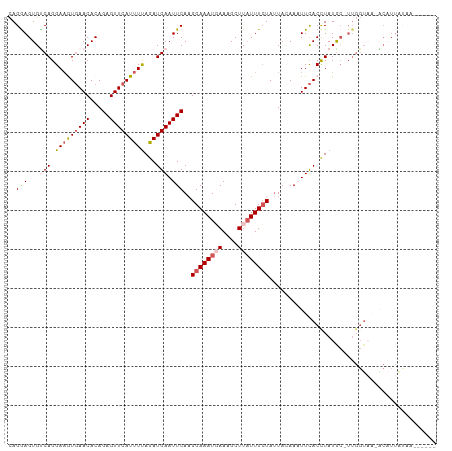
| Location | 3,834,654 – 3,834,760 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Shannon entropy | 0.37906 |
| G+C content | 0.31679 |
| Mean single sequence MFE | -17.50 |
| Consensus MFE | -11.30 |
| Energy contribution | -12.10 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3834654 106 - 21146708 --UUACAUAUAAUGC-UUACCAA-GCAUAGGUGAAUUUUUAAUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUGAAAUGAACUCUGUGUUCACUUCCUGGCACUCCUG --.........((((-(.....)-))))((((((((((......(((((........)))))...))))))))))(((.(((((.....))))).)))............ ( -20.50, z-score = -1.85, R) >droSim1.chr2R 2531036 110 - 19596830 UUAUAAUGGCAAGGUAAAACCAAGGCAUAGGUAAAUUUGUAUUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUAAUUUCCUGACACUCCUG .........(((((.(((...((((((((((....))))).(((....))))))))..))))))))..(((((.....)))))....((((((......))))))..... ( -16.20, z-score = 0.34, R) >droSec1.super_1 1471613 103 - 14215200 ------UUAUAAUGC-UUACCAAGGCAUAGGUGAAUUUGUAAUAGAAAUAAGCUUUAAUUUCCAUGAAUUCAUCUAAAAUGAACUCUGUGUUCAUUUCCUGACACUCCUG ------......(((-((....)))))((((((((((..(....(((((........))))).)..))))))))))((((((((.....))))))))............. ( -23.30, z-score = -3.07, R) >droYak2.chr2L 16491588 102 - 22324452 ------UUAUAAUGU-UUACCAA-GUAUAAGUGAAAUUGUAAUACAAAAAAGCGUUAAUUUCCUUGAAUUCAUUUAAAAUGAACUCUGUGUUCACUUCCUGACACUUCUG ------......(((-(....((-((.((((.(((((((................)))))))))))..............((((.....))))))))...))))...... ( -12.59, z-score = -0.41, R) >droEre2.scaffold_4929 7847467 84 + 26641161 -------------------------CAUAGGUGAAAUUAUAAUAGAAAUAAGCGUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUCAC-UCCUGUCACUUCUA -------------------------...((((((..........(((...((.((((((((...((....))....)))))))).))...)))..-.....))))))... ( -14.93, z-score = -1.94, R) >consensus ______UUAUAAUGC_UUACCAA_GCAUAGGUGAAUUUGUAAUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUCACUUCCUGACACUCCUG ...........................((((((((((((.....(((((........)))))..))))))))))))...(((((.....)))))................ (-11.30 = -12.10 + 0.80)

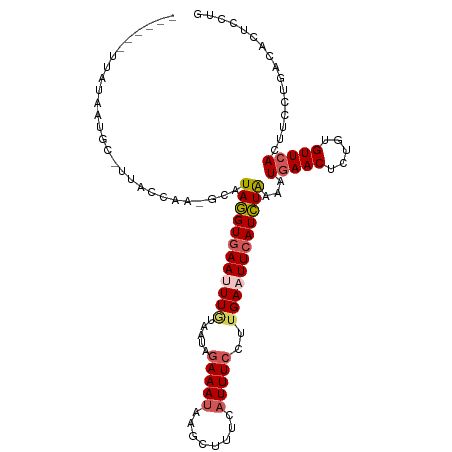
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:54 2011