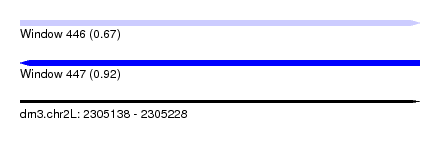
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,305,138 – 2,305,228 |
| Length | 90 |
| Max. P | 0.917251 |

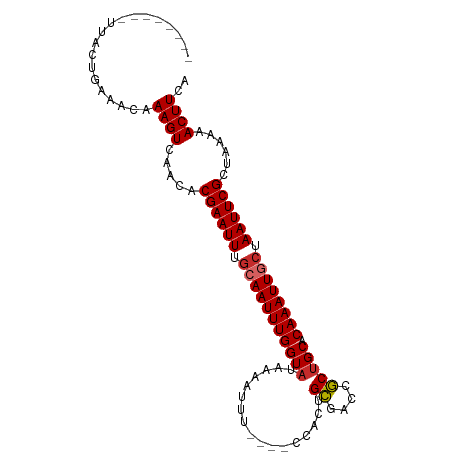
| Location | 2,305,138 – 2,305,228 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.99 |
| Shannon entropy | 0.19551 |
| G+C content | 0.35891 |
| Mean single sequence MFE | -18.39 |
| Consensus MFE | -13.87 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2305138 90 + 23011544 --------UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU----CCACUGCGACCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA --------...........((((.....((((((.(((((((((((........----.....((....))))).)))))))).))))))......)))).. ( -16.41, z-score = -2.02, R) >droEre2.scaffold_4929 2338809 90 + 26641161 --------UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU----CCACUGCGACCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA --------...........((((.....((((((.(((((((((((........----.....((....))))).)))))))).))))))......)))).. ( -16.41, z-score = -2.02, R) >droYak2.chr2L 2283544 90 + 22324452 --------UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAUUUU----CCGCUGCGACCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA --------...........((((.....((((((.(((((((((..........----))((.((....)).))..))))))).))))))......)))).. ( -17.90, z-score = -1.99, R) >droSec1.super_5 476051 90 + 5866729 --------UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU----CCACUGCGACCGCUGCACAAAUUUCUAAUUCGCUAAAAACUUCA --------...........((((.....((((((.(.(((((((((........----.....((....))))).)))))).).))))))......)))).. ( -9.81, z-score = 0.06, R) >droSim1.chr2L 2262434 90 + 22036055 --------UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU----CCACUGCGACCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA --------...........((((.....((((((.(((((((((((........----.....((....))))).)))))))).))))))......)))).. ( -16.41, z-score = -2.02, R) >droAna3.scaffold_12916 10022098 90 - 16180835 --------UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU----CCACUGUGACCACUGCACAAAUUGCUAAUUCGCUAAAAACUUCA --------...........((((.....((((((.(((((((((((........----.............))).)))))))).))))))......)))).. ( -15.30, z-score = -1.89, R) >dp4.chr4_group3 7573970 94 - 11692001 --------UUAGUGAAACAAAGUAAACACGAAUUUACAAUUUGGUAGAAAAUUUUGCAGUGCGGCGACCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA --------((((((((....(((((...(((((.....)))))(((((....))))).(((((((....)))))))...)))))..))))))))........ ( -25.40, z-score = -2.98, R) >droPer1.super_1 4690076 93 - 10282868 --------UUAGUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU-GCAGUGCGGCGACCGCUGCACAAAUUUCUAAUUCGCUAAAAACUUCA --------((((((((..................(((((.((.......)).))-)))(((((((....)))))))..........))))))))........ ( -24.90, z-score = -2.98, R) >droWil1.scaffold_180708 2270980 97 + 12563649 CUGAAAGAGACUUGAGACAAAGUCAACACGAAUUUGCAAUUUGGUUUAAAUUUC-----CAUGGCGGCCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA ..(((...(((((......)))))....((((((.((((((((((.........-----...(((....))))).)))))))).))))))........))). ( -23.00, z-score = -1.65, R) >consensus ________UUACUGAAACAAAGUCAACACGAAUUUGCAAUUUGGUAUAAAAUUU____CCACUGCGACCGCUGCACAAAUUGCUAAUUCGCUAAAAACUUCA ...................((((.....((((((.(((((((((((.................((....))))).)))))))).))))))......)))).. (-13.87 = -14.12 + 0.25)

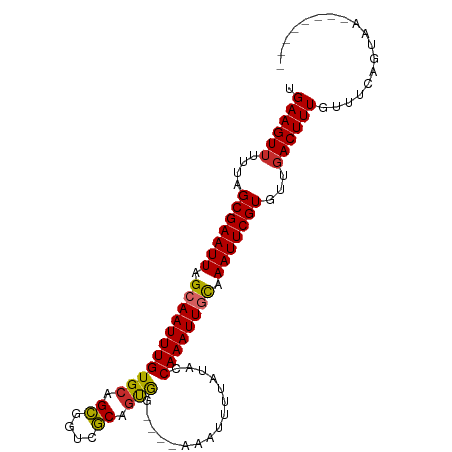
| Location | 2,305,138 – 2,305,228 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.99 |
| Shannon entropy | 0.19551 |
| G+C content | 0.35891 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.51 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2305138 90 - 23011544 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGUCGCAGUGG----AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA-------- .((((((....(((((((.((((((((....((....))(((..----..)))......)))))))).)))))))...))))))..........-------- ( -23.60, z-score = -1.99, R) >droEre2.scaffold_4929 2338809 90 - 26641161 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGUCGCAGUGG----AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA-------- .((((((....(((((((.((((((((....((....))(((..----..)))......)))))))).)))))))...))))))..........-------- ( -23.60, z-score = -1.99, R) >droYak2.chr2L 2283544 90 - 22324452 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGUCGCAGCGG----AAAAUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA-------- .((((((....(((((((.(((((((((((.((....)).))).----...........)))))))).)))))))...))))))..........-------- ( -24.30, z-score = -1.90, R) >droSec1.super_5 476051 90 - 5866729 UGAAGUUUUUAGCGAAUUAGAAAUUUGUGCAGCGGUCGCAGUGG----AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA-------- .((((((....(((((((.(.((((((....((....))(((..----..)))......)))))).).)))))))...))))))..........-------- ( -17.00, z-score = 0.08, R) >droSim1.chr2L 2262434 90 - 22036055 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGUCGCAGUGG----AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA-------- .((((((....(((((((.((((((((....((....))(((..----..)))......)))))))).)))))))...))))))..........-------- ( -23.60, z-score = -1.99, R) >droAna3.scaffold_12916 10022098 90 + 16180835 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGUGGUCACAGUGG----AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA-------- .((((((....(((((((.(((((((((.((.((....)).)).----)..........)))))))).)))))))...))))))..........-------- ( -23.40, z-score = -2.36, R) >dp4.chr4_group3 7573970 94 + 11692001 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGUCGCCGCACUGCAAAAUUUUCUACCAAAUUGUAAAUUCGUGUUUACUUUGUUUCACUAA-------- .(((((.....(((((((.((((((((((((((((...))))..))))...........)))))))).)))))))....)))))..........-------- ( -25.60, z-score = -2.77, R) >droPer1.super_1 4690076 93 + 10282868 UGAAGUUUUUAGCGAAUUAGAAAUUUGUGCAGCGGUCGCCGCACUGC-AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCACUAA-------- .((((((....(((((((........((((.((....)).))))(((-((.(((......)))))))))))))))...))))))..........-------- ( -23.00, z-score = -1.50, R) >droWil1.scaffold_180708 2270980 97 - 12563649 UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGCCGCCAUG-----GAAAUUUAAACCAAAUUGCAAAUUCGUGUUGACUUUGUCUCAAGUCUCUUUCAG .((((((....(((((((.((((((((........((.....)-----)..........)))))))).)))))))...)))))).................. ( -22.17, z-score = -0.84, R) >consensus UGAAGUUUUUAGCGAAUUAGCAAUUUGUGCAGCGGUCGCAGUGG____AAAUUUUAUACCAAAUUGCAAAUUCGUGUUGACUUUGUUUCAGUAA________ .((((((....(((((((.(((((((((((.((....)).)))................)))))))).)))))))...)))))).................. (-20.76 = -20.51 + -0.24)

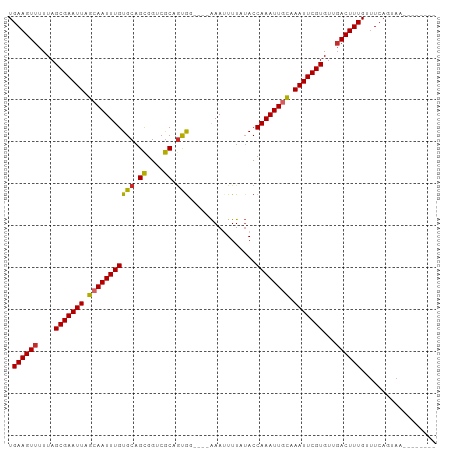
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:51 2011