| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,793,394 – 3,793,487 |
| Length | 93 |
| Max. P | 0.972365 |

| Location | 3,793,394 – 3,793,487 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 62.13 |
| Shannon entropy | 0.74609 |
| G+C content | 0.64625 |
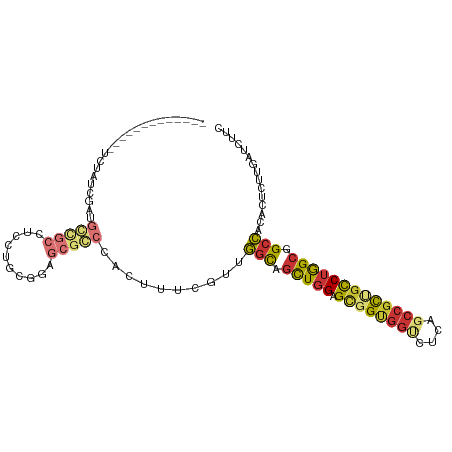
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.54 |
| Covariance contribution | 0.53 |
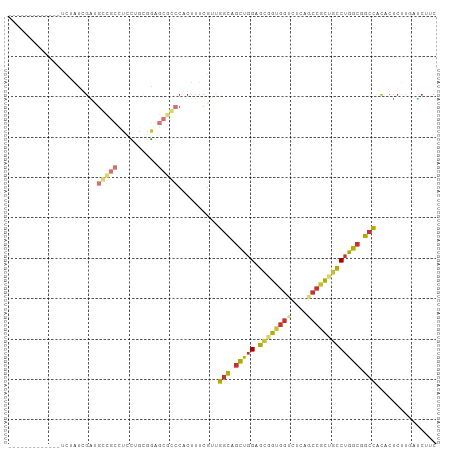
| Combinations/Pair | 1.59 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
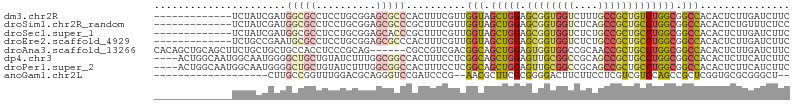
>dm3.chr2R 3793394 93 - 21146708 -------------UCUAUCGAUGGCGCCUCCUGCGGAGCGCCCACUUUCGUUGGUAGCUGGAGCGGUGGUCUUUGCCGCUGUCUGGCGGCCACACUCUUGAUCUUC -------------...(((((.(((((.(((...)))))))).......((((((.((..((.(((((((....)))))))))..)).)))).))..))))).... ( -36.80, z-score = -1.46, R) >droSim1.chr2R_random 1031816 93 - 2996586 -------------UCUAUCGAUGGCGCCUCCUGCGGAGCGCCCGCUUUCGUUGGUAGCUGGAGCGGUGGUCUCAGCCGCUGCCUGGCGGCCACACUCUGUUUCUCC -------------.....(((.(((((.(((...)))))))).....))).((((.((..(.((((((((....)))))))))..)).)))).............. ( -37.60, z-score = -0.55, R) >droSec1.super_1 1429825 93 - 14215200 -------------UCUAUCGAUGGCGCCUCCUGCGGAGCACCCGCUUUCGUUGGUAGCUGGAGCGGUGGUCUCUGCCGCUGCCUGGCGGCCACACUCUUGAUCUUC -------------...((((((((((((....(((((((....)).))))).))).((..(.((((((((....)))))))))..)).)))).....))))).... ( -35.90, z-score = -0.76, R) >droEre2.scaffold_4929 7807238 93 + 26641161 -------------UCUGCCGAAUGCGCCUCCUGCGGAGCGCCCACUUUCGUUGGUAGCUGGAGCGGUGGUCUCUGCCGCUGCCUGGCGGCCACACUCUUGAUCUUC -------------.....((((.((((.(((...))))))).....)))).((((.((..(.((((((((....)))))))))..)).)))).............. ( -35.20, z-score = -0.50, R) >droAna3.scaffold_13266 16115876 100 - 19884421 CACAGCUGCAGCUUCUGCUGCUGCCACCUCCCGCAG------CGCCGUCGACGGCAGCUGGAGUGGUGGCCGCAACCGCUGCCUGGCGGCCACACUCUUGAUCUUC ..(((((((((...)))).(((((........))))------)((((....)))))))))(((((.(((((((............))))))))))))......... ( -46.50, z-score = -1.89, R) >dp4.chr3 6371010 102 + 19779522 ----ACUGGCAAUGGCAAUGGGGCUGCUGUAUCUUUGGCGGCCACUUUCCUCGGCAGCUGGAGUUGCGGCCGCAGCCGCUGCCUGGCGGCCACACUCUUCAUCUUC ----..((((...((((.(((.((.((((((.(((..((.(((.........))).))..))).)))))).))..))).)))).....)))).............. ( -42.50, z-score = -0.97, R) >droPer1.super_2 8213870 102 - 9036312 ----ACUGGCAAUGGCAAUGGGGCUGCUGUAUCUUUGGCGGCCACUUUCCUCGGCAGCUGGAGUUGCGGCCGCAGCCGCUGCCUGGCGGCCACACUCUUCAUCUUC ----..((((...((((.(((.((.((((((.(((..((.(((.........))).))..))).)))))).))..))).)))).....)))).............. ( -42.50, z-score = -0.97, R) >anoGam1.chr2L 38347433 83 - 48795086 -------------------CUUGCCGGUUUGGACGCAGGGUCCGAUCCCG--AACGCUUCGCGGGGACUUCUUCCUCGUCGUCCAGCCGCUCGGUGCGCGGGCU-- -------------------...(((((((.(((((..(((......))).--........(((((((.....))))))))))))))))((.....))...))).-- ( -33.00, z-score = 0.49, R) >consensus _____________UCUAUCGAUGCCGCCUCCUGCGGAGCGCCCACUUUCGUUGGCAGCUGGAGCGGUGGUCUCAGCCGCUGCCUGGCGGCCACACUCUUGAUCUUC ......................(((((.(......).)))))..........(((.(((((.((((((((....))))))))))))).)))............... (-22.00 = -22.54 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:49 2011