| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,302,069 – 2,302,138 |
| Length | 69 |
| Max. P | 0.683763 |

| Location | 2,302,069 – 2,302,138 |
|---|---|
| Length | 69 |
| Sequences | 7 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 60.93 |
| Shannon entropy | 0.72898 |
| G+C content | 0.44729 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -4.69 |
| Energy contribution | -5.39 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
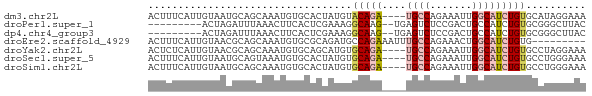
>dm3.chr2L 2302069 69 + 23011544 ACUUUCAUUGUAAUGCAGCAAAUGUGCACUAUGUACAGA----UGCCAGAAAUUGGCAUCUGUGCAUAGGAAA ...(((.......((((.(....)))))(((((((((((----((((((...)))))))))))))))))))). ( -30.50, z-score = -5.56, R) >droPer1.super_1 4685935 62 - 10282868 ---------ACUAGAUUUAAACUUCACUCGAAAGGCAAG--UGAGUCUCCGACUGCCAUCUGUGCGGGCUUAC ---------...((((((..((((...((....)).)))--)))))))....(((((....).))))...... ( -10.40, z-score = 0.91, R) >dp4.chr4_group3 7569805 62 - 11692001 ---------ACUAGAUUUAAACUUCACUCGAAAGGCAAG--UGAGUCUCCGACUGCCAUCUGUGCGGGCUUAC ---------...((((((..((((...((....)).)))--)))))))....(((((....).))))...... ( -10.40, z-score = 0.91, R) >droEre2.scaffold_4929 2335709 64 + 26641161 ACUUUCAUUGUAACGCAGCAAAUGUGCGCAGAUGCCAGAAAUUUGCCAGAAACUGGCAUCUGUG--------- .............((((.....))))((((((((((((..............))))))))))))--------- ( -21.44, z-score = -2.59, R) >droYak2.chr2L 2280405 69 + 22324452 ACUCUCAUUGUAACGCAGCAAAUGUGCAGCAUGUGCAGA----UGCCAGAAAUUGGCAUCUGUGCCUAGGAAA ..(((.........((.(((....))).))..(..((((----((((((...))))))))))..)...))).. ( -23.40, z-score = -1.76, R) >droSec1.super_5 472907 69 + 5866729 ACUUUCAUUGUAAUGCAGUAAAUGUGCACUAUGUGCAGA----UGCCAGAAAUUGGCAUCUGUGCCUGGGAAA ...(((.......((((.......))))(((.(..((((----((((((...))))))))))..).)))))). ( -24.80, z-score = -3.06, R) >droSim1.chr2L 2259311 69 + 22036055 ACUUUCAUUGUAAUGCAGCAAAUGUGCACUAUGUGCAGA----UGCCAGAAAUUGGCAUCUGUGCCUGGGAAA ...(((.......((((.(....)))))(((.(..((((----((((((...))))))))))..).)))))). ( -25.10, z-score = -2.65, R) >consensus ACUUUCAUUGUAAUGCAGCAAAUGUGCACUAUGUGCAGA____UGCCAGAAAUUGGCAUCUGUGCCUGGGAAA ..................................(((((....((((.......))))))))).......... ( -4.69 = -5.39 + 0.70)


| Location | 2,302,069 – 2,302,138 |
|---|---|
| Length | 69 |
| Sequences | 7 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 60.93 |
| Shannon entropy | 0.72898 |
| G+C content | 0.44729 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -4.72 |
| Energy contribution | -5.56 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2302069 69 - 23011544 UUUCCUAUGCACAGAUGCCAAUUUCUGGCA----UCUGUACAUAGUGCACAUUUGCUGCAUUACAAUGAAAGU (((((((((.((((((((((.....)))))----))))).)))))(((((....).)))).......)))).. ( -25.30, z-score = -4.37, R) >droPer1.super_1 4685935 62 + 10282868 GUAAGCCCGCACAGAUGGCAGUCGGAGACUCA--CUUGCCUUUCGAGUGAAGUUUAAAUCUAGU--------- .(((((...(((.((.((((((.(....)..)--).))))..))..)))..)))))........--------- ( -14.70, z-score = -0.56, R) >dp4.chr4_group3 7569805 62 + 11692001 GUAAGCCCGCACAGAUGGCAGUCGGAGACUCA--CUUGCCUUUCGAGUGAAGUUUAAAUCUAGU--------- .(((((...(((.((.((((((.(....)..)--).))))..))..)))..)))))........--------- ( -14.70, z-score = -0.56, R) >droEre2.scaffold_4929 2335709 64 - 26641161 ---------CACAGAUGCCAGUUUCUGGCAAAUUUCUGGCAUCUGCGCACAUUUGCUGCGUUACAAUGAAAGU ---------..((((((((((..............))))))))))(((((....).))))............. ( -19.54, z-score = -1.68, R) >droYak2.chr2L 2280405 69 - 22324452 UUUCCUAGGCACAGAUGCCAAUUUCUGGCA----UCUGCACAUGCUGCACAUUUGCUGCGUUACAAUGAGAGU ((((.......(((((((((.....)))))----))))...((((.(((....))).))))......)))).. ( -21.00, z-score = -1.39, R) >droSec1.super_5 472907 69 - 5866729 UUUCCCAGGCACAGAUGCCAAUUUCUGGCA----UCUGCACAUAGUGCACAUUUACUGCAUUACAAUGAAAGU ((((.......(((((((((.....)))))----))))....(((((((.......)))))))....)))).. ( -20.60, z-score = -2.94, R) >droSim1.chr2L 2259311 69 - 22036055 UUUCCCAGGCACAGAUGCCAAUUUCUGGCA----UCUGCACAUAGUGCACAUUUGCUGCAUUACAAUGAAAGU ((((.......(((((((((.....)))))----))))....((((((((....).)))))))....)))).. ( -20.90, z-score = -2.16, R) >consensus UUUCCCAGGCACAGAUGCCAAUUUCUGGCA____UCUGCACAUAGUGCACAUUUGCUGCAUUACAAUGAAAGU ...........(((((((((.....)))))....))))......(((((.......)))))............ ( -4.72 = -5.56 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:49 2011