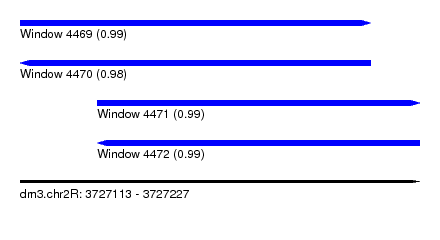
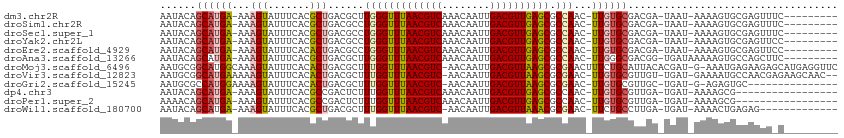
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,727,113 – 3,727,227 |
| Length | 114 |
| Max. P | 0.994869 |

| Location | 3,727,113 – 3,727,213 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.44 |
| Shannon entropy | 0.37605 |
| G+C content | 0.42670 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.21 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
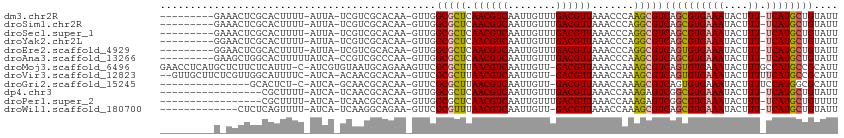
>dm3.chr2R 3727113 100 + 21146708 ---------GAAACUCGCACUUUU-AUUA-UCGUCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAAGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUU ---------......(((......-.(((-.(((((.((((-.((((((.....)))))))))).))))).)))......))).((((((((((....))-)))))))).... ( -30.72, z-score = -3.65, R) >droSim1.chr2R 2444045 100 + 19596830 ---------GAAACUCGCACUUUU-AUUA-UCGUCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUU ---------......(((.((...-.(((-.(((((.((((-.((((((.....)))))))))).))))).)))....))))).((((((((((....))-)))))))).... ( -32.60, z-score = -3.96, R) >droSec1.super_1 1374082 100 + 14215200 ---------GAAACUCGCACUUUU-AUUA-UCGUCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUU ---------......(((.((...-.(((-.(((((.((((-.((((((.....)))))))))).))))).)))....))))).((((((((((....))-)))))))).... ( -32.60, z-score = -3.96, R) >droYak2.chr2L 16391325 100 + 22324452 ---------GGAACUCGCACUUUU-AUUA-UCGUCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUU ---------......(((.((...-.(((-.(((((.((((-.((((((.....)))))))))).))))).)))....))))).((((((((((....))-)))))))).... ( -32.60, z-score = -3.66, R) >droEre2.scaffold_4929 7750577 100 - 26641161 ---------GGAACUCGCACUUUU-AUUA-UCGUCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGUGUGAAAUACUUU-UCAUGCUGUAUU ---------......(((.((...-.(((-.(((((.((((-.((((((.....)))))))))).))))).)))....))))).((((((((((....))-)))))))).... ( -30.00, z-score = -2.88, R) >droAna3.scaffold_13266 16062214 101 + 19884421 ---------GAAGCUGGCACUUUUUAUCA-CCGUCGCCCAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAAGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUU ---------...(((((............-..(.((((...-...)))))..((((((((....))))))))....)).)))..((((((((((....))-)))))))).... ( -32.70, z-score = -3.59, R) >droMoj3.scaffold_6496 687192 110 + 26866924 GAACCUCAUGCUCUUCUCAUUU-C-AUCGUGUAAUGCAGAAAGUUCGCGCUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUGCCAUGCCGCAUU ......................-.-...(((..(((((((...((((((((((((((((.....)-)))))))............))))))))....)))).)))..)))... ( -24.10, z-score = -0.81, R) >droVir3.scaffold_12823 1251705 107 - 2474545 --GUUGCUUCUCGUUGGCAUUUUC-AUCA-ACAACGCACAA-GUUCGCGCUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUUUCAUGCCGCAUU --((.((((..(((((........-....-.)))))...))-))..))..(((((((((.....)-))))))))......(((...((((((((.....)))))))))))... ( -26.12, z-score = -1.39, R) >droGri2.scaffold_15245 6002570 93 + 18325388 ---------------GCACUCU-C-AUCA-GCAACGCACAA-GUUCGCGCUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUUCCAUGGCGCAUU ---------------.......-.-....-(((((......-))).))..(((((((((.....)-))))))))......((((((.((.((((....))))))))))))... ( -25.40, z-score = -2.22, R) >dp4.chr3 6317083 92 - 19779522 -----------------CGCUUUU-AUCA-UCAACGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGAGUCGGCGUGAAAUACUUU-UCAUGCUGUAUU -----------------.((((((-....-(((((......-))))).....((((((((....))))))))......))))))((((((((((....))-)))))))).... ( -28.30, z-score = -3.33, R) >droPer1.super_2 8158852 92 + 9036312 -----------------CGCUUUU-AUCA-UCAACGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGAGUCGGCGUGAAAUACUUU-UCAUGCUGUUUU -----------------.((((((-....-(((((......-))))).....((((((((....))))))))......))))))((((((((((....))-)))))))).... ( -28.30, z-score = -3.24, R) >droWil1.scaffold_180700 1187819 95 - 6630534 -------------CUCUCAGUUUU-AUCA-UCAAGGCAGAA-GUUCGCGUUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUU -------------......(((((-....-.....((....-....))(((((((((((.....)-)))))))))).)))))..((((((((((....))-)))))))).... ( -29.90, z-score = -4.62, R) >consensus _________G_AACUCGCACUUUU_AUCA_UCAACGCACAA_GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGCGUCAGCGUGAAAUACUUU_UCAUGCUGUAUU ..............................................(((((.((((((........)))))).......)))))((((((((.(.....).)))))))).... (-15.16 = -15.21 + 0.05)
| Location | 3,727,113 – 3,727,213 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Shannon entropy | 0.37605 |
| G+C content | 0.42670 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.21 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3727113 100 - 21146708 AAUACAGCAUGA-AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGACGA-UAAU-AAAAGUGCGAGUUUC--------- ....((((.(((-((....))))).)))).....(((((((((((((((....)))))))))))).)))((-(((..(.....-....-....)..)))))...--------- ( -34.92, z-score = -3.95, R) >droSim1.chr2R 2444045 100 - 19596830 AAUACAGCAUGA-AAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGACGA-UAAU-AAAAGUGCGAGUUUC--------- ....((((.(((-((....))))).))))......((((((((((((((....)))))))))))).))(((-(((..(.....-....-....)..))))))..--------- ( -34.92, z-score = -4.02, R) >droSec1.super_1 1374082 100 - 14215200 AAUACAGCAUGA-AAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGACGA-UAAU-AAAAGUGCGAGUUUC--------- ....((((.(((-((....))))).))))......((((((((((((((....)))))))))))).))(((-(((..(.....-....-....)..))))))..--------- ( -34.92, z-score = -4.02, R) >droYak2.chr2L 16391325 100 - 22324452 AAUACAGCAUGA-AAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGACGA-UAAU-AAAAGUGCGAGUUCC--------- ....((((.(((-((....))))).))))......((((((((((((((....)))))))))))).))(((-(((..(.....-....-....)..))))))..--------- ( -34.92, z-score = -4.11, R) >droEre2.scaffold_4929 7750577 100 + 26641161 AAUACAGCAUGA-AAAGUAUUUCACACUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGACGA-UAAU-AAAAGUGCGAGUUCC--------- ....(((..(((-((....)))))..)))......((((((((((((((....)))))))))))).))(((-(((..(.....-....-....)..))))))..--------- ( -31.02, z-score = -3.20, R) >droAna3.scaffold_13266 16062214 101 - 19884421 AAUACAGCAUGA-AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGGGCGACGG-UGAUAAAAAGUGCCAGCUUC--------- ....((((.(((-((....))))).))))..((((((((((((((((((....)))))))))))).)))..-...(((.((..-(.....)..))))))))...--------- ( -34.90, z-score = -3.37, R) >droMoj3.scaffold_6496 687192 110 - 26866924 AAUGCGGCAUGGCAAAGUAUUUCACACUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAGCGCGAACUUUCUGCAUUACACGAU-G-AAAUGAGAAGAGCAUGAGGUUC .((((.((....(((((..((.......))..)))))((((((((((-(.....)))))))))))))....(((((.((((.(.....-)-.))))))))).))))....... ( -30.00, z-score = -1.58, R) >droVir3.scaffold_12823 1251705 107 + 2474545 AAUGCGGCAUGAAAAAGUAUUUCACACUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAGCGCGAAC-UUGUGCGUUGU-UGAU-GAAAAUGCCAACGAGAAGCAAC-- .....(((((.........(((((((..(((((((((((((((((((-(.....))))))))))).)))).-....)))))..-)).)-))))))))).............-- ( -31.10, z-score = -1.79, R) >droGri2.scaffold_15245 6002570 93 - 18325388 AAUGCGCCAUGGAAAAGUAUUUCACACUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAGCGCGAAC-UUGUGCGUUGC-UGAU-G-AGAGUGC--------------- ...((((..(....)....(((((((..(((((((((((((((((((-(.....))))))))))).)))).-....)))))..-)).)-)-)))))))--------------- ( -27.30, z-score = -1.14, R) >dp4.chr3 6317083 92 + 19779522 AAUACAGCAUGA-AAAGUAUUUCACGCCGACUCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGUUGA-UGAU-AAAAGCG----------------- ......((..((-((....))))(((((((....(((((((((((((((....))))))))))..))))).-))).))))...-....-....)).----------------- ( -23.70, z-score = -1.48, R) >droPer1.super_2 8158852 92 - 9036312 AAAACAGCAUGA-AAAGUAUUUCACGCCGACUCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGUUGA-UGAU-AAAAGCG----------------- ......((..((-((....))))(((((((....(((((((((((((((....))))))))))..))))).-))).))))...-....-....)).----------------- ( -23.70, z-score = -1.45, R) >droWil1.scaffold_180700 1187819 95 + 6630534 AAUACAGCAUGA-AAAGUAUUUCACGCUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAACGCGAAC-UUCUGCCUUGA-UGAU-AAAACUGAGAG------------- ....((((.(((-((....))))).)))).(((....((((((((((-(.....))))))))))))))...-...........-....-...........------------- ( -25.10, z-score = -3.02, R) >consensus AAUACAGCAUGA_AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC_UUGUGCGACGA_UGAU_AAAAGUGCGAGUU_C_________ ......((((((...(((.......)))......(((((((((((((........)))))))))).)))...))))))................................... (-17.53 = -17.21 + -0.32)
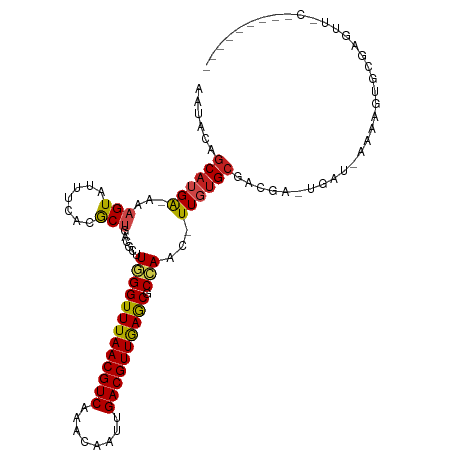
| Location | 3,727,135 – 3,727,227 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Shannon entropy | 0.40119 |
| G+C content | 0.46821 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.03 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3727135 92 + 21146708 UCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAAGCGUCAGCGUGAAAUACUUUU-CAUGCUGUAUUACCCGAGCAGAGUG-------- ..((....-....))((((((((((((....))))))))............((((((((((....)))-)))))))........))))......-------- ( -30.50, z-score = -3.36, R) >droSim1.chr2R 2444067 92 + 19596830 UCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGCGUGAAAUACUUUU-CAUGCUGUAUUACCCGGGCAGAGUG-------- ..((....-....))((((((((((((....))))))))...(((.((...((((((((((....)))-))))))).....)).)))..)))).-------- ( -35.10, z-score = -3.94, R) >droSec1.super_1 1374104 92 + 14215200 UCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGCGUGAAAUACUUUU-CAUGCUGUAUUACCCGGGCAGAGUG-------- ..((....-....))((((((((((((....))))))))...(((.((...((((((((((....)))-))))))).....)).)))..)))).-------- ( -35.10, z-score = -3.94, R) >droYak2.chr2L 16391347 97 + 22324452 UCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGCGUGAAAUACUUUU-CAUGCUGUAUUACCCGAACCGAGCGGAGUG--- ........-.....(((((((((((((....)))))))).......((.((((((((((((....)))-)))))))....)))).....))))).....--- ( -33.30, z-score = -2.90, R) >droEre2.scaffold_4929 7750599 92 - 26641161 UCGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAGGCGUCAGUGUGAAAUACUUUU-CAUGCUGUAUUACCCGAGCAGAGUG-------- ..((....-....))((((((((((((....)))))))).......((.((((((((((((....)))-)))))))....))))))))......-------- ( -30.00, z-score = -2.88, R) >droAna3.scaffold_13266 16062237 100 + 19884421 UCGCCCAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAAGCGUCAGCGUGAAAUACUUUU-CAUGCUGUAUUACCCAGUGGGGGCGGGUGGGUG .((((...-...))))...((((((((....))))))))..(((((..(((((((((((((....)))-))))))).....(((....))))))..))))). ( -42.40, z-score = -3.88, R) >droMoj3.scaffold_6496 687223 101 + 26866924 AUGCAGAAAGUUCGCGCUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUGCCAUGCCGCAUUACUCAUACUCUGCAGACGCACA .((((((......(((((((((((((.....)-)))))))......)))))..((((((.......(((......)))....))))))))))))........ ( -28.30, z-score = -1.72, R) >droVir3.scaffold_12823 1251734 90 - 2474545 ACGCACAA-GUUCGCGCUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUUUCAUGCCGCAUUACCC-GCGGCCCA--------- .(((((..-...(((..(((((((((.....)-))))))))......)))...)))))..............(((((.......-)))))...--------- ( -27.10, z-score = -2.68, R) >droGri2.scaffold_15245 6002585 95 + 18325388 ACGCACAA-GUUCGCGCUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUUCCAUGGCGCAUUACCUUGUUUCCCGCCCA----- ..((((((-(.......(((((((((.....)-))))))))......((((((.((.((((....)))))))))))).....))))).....))...----- ( -25.50, z-score = -2.44, R) >dp4.chr3 6317097 97 - 19779522 ACGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGAGUCGGCGUGAAAUACUUUU-CAUGCUGUAUUACCCCAUGAAAGGAGGAUG--- .(((....-....)))...((((((((....)))))))).........(((((((((((((....)))-))))))).))).((((......)).))...--- ( -28.10, z-score = -2.07, R) >droPer1.super_2 8158866 97 + 9036312 ACGCACAA-GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGAGUCGGCGUGAAAUACUUUU-CAUGCUGUUUUACCCCAUGAAAGGAGGAUG--- .(((....-....)))...((((((((....)))))))).......((((.((((((((((....)))-))))))))))).((((......)).))...--- ( -28.60, z-score = -2.09, R) >droWil1.scaffold_180700 1187837 91 - 6630534 AGGCAGAA-GUUCGCGUUUAACGUCAAUUGUU-GACGUUAAACCAAAGCGUCAGCGUGAAAUACUUUU-CAUGCUGUAUUACCCAACCGAUGUA-------- ..(((...-(((((((((((((((((.....)-))))))))))....))).((((((((((....)))-)))))))........)))...))).-------- ( -30.00, z-score = -4.10, R) >consensus ACGCACAA_GUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGCGUCAGCGUGAAAUACUUUU_CAUGCUGUAUUACCCGAGCAGAGUG________ .............(((((.((((((........)))))).......)))))((((((((((....))).))))))).......................... (-15.07 = -15.03 + -0.03)
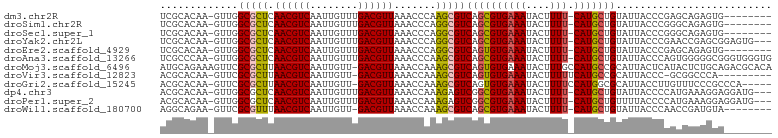
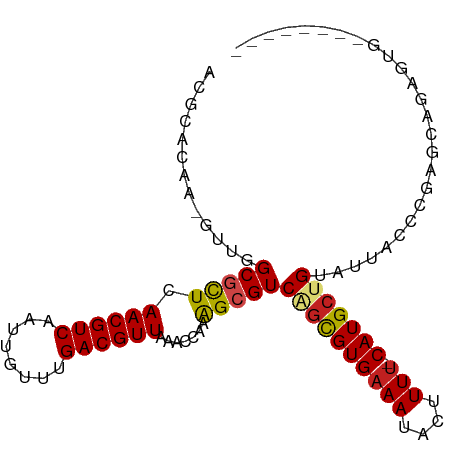
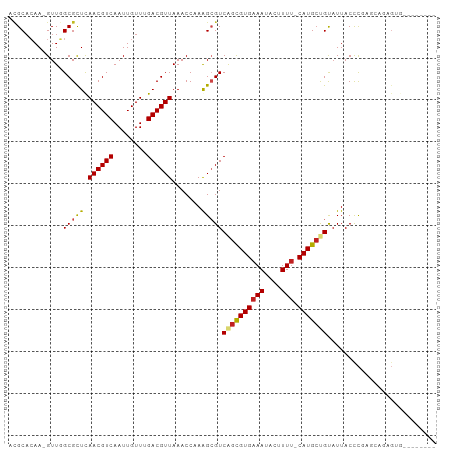
| Location | 3,727,135 – 3,727,227 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Shannon entropy | 0.40119 |
| G+C content | 0.46821 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -17.42 |
| Energy contribution | -16.58 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
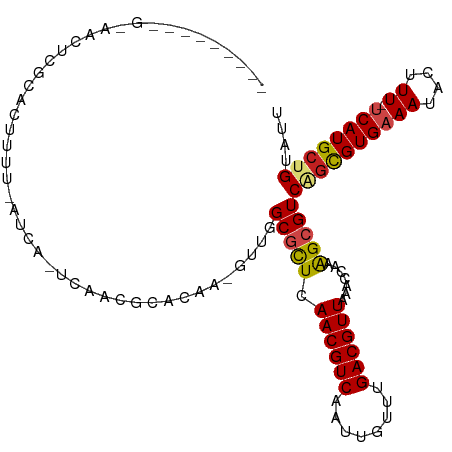
>dm3.chr2R 3727135 92 - 21146708 --------CACUCUGCUCGGGUAAUACAGCAUG-AAAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGA --------.....(((.(((((....((((.((-(((....))))).)))).....(((((((((((((((....)))))))))))).)))))-))).))). ( -35.30, z-score = -4.28, R) >droSim1.chr2R 2444067 92 - 19596830 --------CACUCUGCCCGGGUAAUACAGCAUG-AAAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGA --------......((((((((....((((.((-(((....))))).))))..))))))))((((((((((....))))))))))((((....-..)))).. ( -37.20, z-score = -4.66, R) >droSec1.super_1 1374104 92 - 14215200 --------CACUCUGCCCGGGUAAUACAGCAUG-AAAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGA --------......((((((((....((((.((-(((....))))).))))..))))))))((((((((((....))))))))))((((....-..)))).. ( -37.20, z-score = -4.66, R) >droYak2.chr2L 16391347 97 - 22324452 ---CACUCCGCUCGGUUCGGGUAAUACAGCAUG-AAAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGA ---.....(((.(((((.((((....((((.((-(((....))))).))))..))))((((((((((((((....)))))))))))).)))))-.)).))). ( -35.50, z-score = -3.18, R) >droEre2.scaffold_4929 7750599 92 + 26641161 --------CACUCUGCUCGGGUAAUACAGCAUG-AAAAGUAUUUCACACUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGA --------.....(((.(((((....(((..((-(((....)))))..))).....(((((((((((((((....)))))))))))).)))))-))).))). ( -31.30, z-score = -3.24, R) >droAna3.scaffold_13266 16062237 100 - 19884421 CACCCACCCGCCCCCACUGGGUAAUACAGCAUG-AAAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGGGCGA ........(((((...(..(((....((((.((-(((....))))).))))..)))..)((((((((((((....))))))))))))......-..))))). ( -38.60, z-score = -4.06, R) >droMoj3.scaffold_6496 687223 101 - 26866924 UGUGCGUCUGCAGAGUAUGAGUAAUGCGGCAUGGCAAAGUAUUUCACACUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAGCGCGAACUUUCUGCAU ........((((((((((.....)))).((....(((((..((.......))..)))))((((((((((-(.....)))))))))))))......)))))). ( -31.90, z-score = -1.51, R) >droVir3.scaffold_12823 1251734 90 + 2474545 ---------UGGGCCGC-GGGUAAUGCGGCAUGAAAAAGUAUUUCACACUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAGCGCGAAC-UUGUGCGU ---------...(((((-(.....)))))).(((((.....))))).....((((((((((((((((((-(.....))))))))))).)))).-....)))) ( -30.90, z-score = -2.13, R) >droGri2.scaffold_15245 6002585 95 - 18325388 -----UGGGCGGGAAACAAGGUAAUGCGCCAUGGAAAAGUAUUUCACACUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAGCGCGAAC-UUGUGCGU -----...(((....(((((.....(((.((((((((....)))).)).)).)))((((((((((((((-(.....))))))))))).)))))-)))).))) ( -28.30, z-score = -1.54, R) >dp4.chr3 6317097 97 + 19779522 ---CAUCCUCCUUUCAUGGGGUAAUACAGCAUG-AAAAGUAUUUCACGCCGACUCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGU ---........(((((((..(.....)..))))-)))........(((((((....(((((((((((((((....))))))))))..))))).-))).)))) ( -26.60, z-score = -1.93, R) >droPer1.super_2 8158866 97 - 9036312 ---CAUCCUCCUUUCAUGGGGUAAAACAGCAUG-AAAAGUAUUUCACGCCGACUCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC-UUGUGCGU ---........(((((((..(.....)..))))-)))........(((((((....(((((((((((((((....))))))))))..))))).-))).)))) ( -26.60, z-score = -1.88, R) >droWil1.scaffold_180700 1187837 91 + 6630534 --------UACAUCGGUUGGGUAAUACAGCAUG-AAAAGUAUUUCACGCUGACGCUUUGGUUUAACGUC-AACAAUUGACGUUAAACGCGAAC-UUCUGCCU --------......(((.((((....((((.((-(((....))))).)))).(((....((((((((((-(.....)))))))))))))).))-))..))). ( -30.30, z-score = -4.14, R) >consensus ________CACUCUGAUCGGGUAAUACAGCAUG_AAAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAAC_UUGUGCGA ................(((((((((((...........)))))((.....)).))))))((((((((((........))))))))))((((....)).)).. (-17.42 = -16.58 + -0.84)

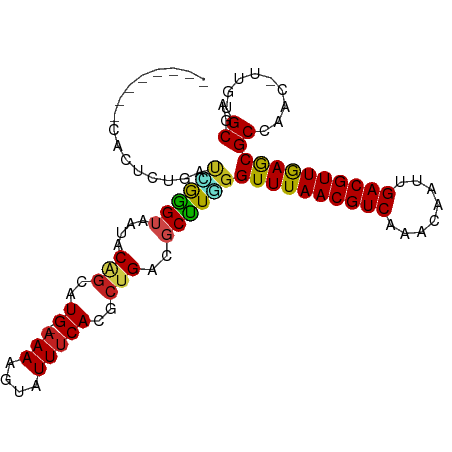
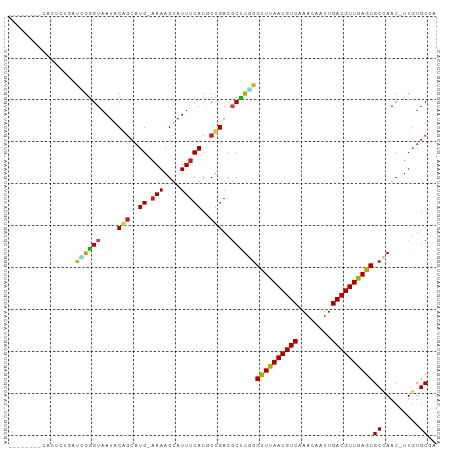
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:39 2011