
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,716,509 – 3,716,593 |
| Length | 84 |
| Max. P | 0.999186 |

| Location | 3,716,509 – 3,716,593 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 72.54 |
| Shannon entropy | 0.54155 |
| G+C content | 0.49435 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -16.57 |
| Energy contribution | -18.87 |
| Covariance contribution | 2.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
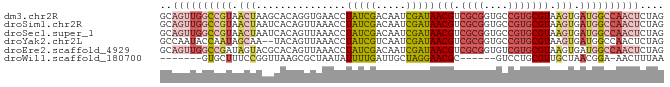
>dm3.chr2R 3716509 84 + 21146708 GCAGUUGGCCGUAACUAAGCACAGGUGAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAG ..((((((((((.(((..((((..((((...(((((.....)))))...))))(....)))))...))).)))))))))).... ( -34.50, z-score = -3.78, R) >droSim1.chr2R 2436467 84 + 19596830 GCAGUUGGCCGUAACUAAUCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAG ..((((((((((.(((...............(((((.....)))))(((.((((....))))))).))).)))))))))).... ( -30.90, z-score = -3.83, R) >droSec1.super_1 1366579 84 + 14215200 GCAGUUGGCCGUAACUAAUCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAG ..((((((((((.(((...............(((((.....)))))(((.((((....))))))).))).)))))))))).... ( -30.90, z-score = -3.83, R) >droYak2.chr2L 16382864 82 + 22324452 GCCAAUACCAAUAGCAA--UACAGUUAAACCUAUCGUCAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAG ((((.(((...((((..--....))))....(((((.....)))))(((.((((....)))))))..))).))))......... ( -20.00, z-score = -1.55, R) >droEre2.scaffold_4929 7742860 84 - 26641161 GCAGUUGGCCGAUAGUACGCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGUCGUGCGUAAGUGAUGGCCAACUCUAG ..(((((((((.((.(((((((((......))...((((..((((...))))...)))))))))))..)).))))))))).... ( -31.60, z-score = -3.21, R) >droWil1.scaffold_180700 5427805 70 + 6630534 -------GUGCUUUCCGGUUAAGCGCUAAUAUUUUGAUUGCUAGGAACGC------GUCCUGCGUUGCUAACGGA-AACUUUAA -------((((((.......)))))).............(.(((.(((((------.....))))).))).)(..-..)..... ( -17.10, z-score = -0.60, R) >consensus GCAGUUGGCCGUAACUAAUCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAG ..((((((((((.(((...............(((((.....)))))(((.((((....))))))).))).)))))))))).... (-16.57 = -18.87 + 2.30)

| Location | 3,716,509 – 3,716,593 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 72.54 |
| Shannon entropy | 0.54155 |
| G+C content | 0.49435 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -12.39 |
| Energy contribution | -14.22 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
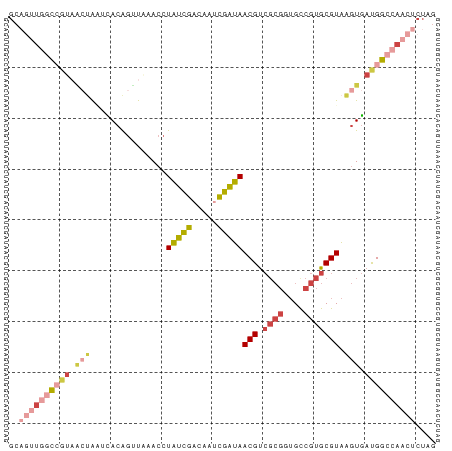
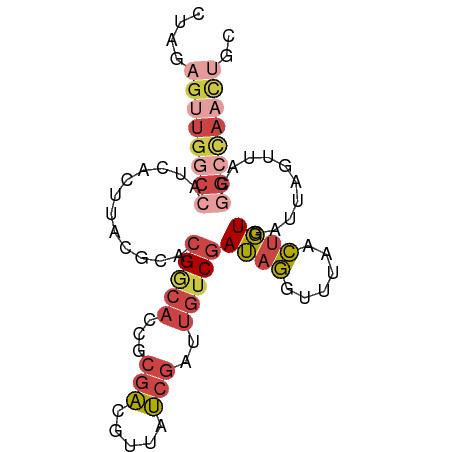
>dm3.chr2R 3716509 84 - 21146708 CUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUCACCUGUGCUUAGUUACGGCCAACUGC ....((((((((...(((...((((((.(((.(((((........)))))...)))....))))))..)))...)))))))).. ( -33.30, z-score = -3.78, R) >droSim1.chr2R 2436467 84 - 19596830 CUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGAUUAGUUACGGCCAACUGC ....((((((((...(((....(((((.(((.(((((........)))))...)))....)))))...)))...)))))))).. ( -30.60, z-score = -3.41, R) >droSec1.super_1 1366579 84 - 14215200 CUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGAUUAGUUACGGCCAACUGC ....((((((((...(((....(((((.(((.(((((........)))))...)))....)))))...)))...)))))))).. ( -30.60, z-score = -3.41, R) >droYak2.chr2L 16382864 82 - 22324452 CUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGACGAUAGGUUUAACUGUA--UUGCUAUUGGUAUUGGC .........((((..(((...(((..(((....(((.((((((.....)))))))))....))).--.)))....)))..)))) ( -17.80, z-score = 0.47, R) >droEre2.scaffold_4929 7742860 84 + 26641161 CUAGAGUUGGCCAUCACUUACGCACGACACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGCGUACUAUCGGCCAACUGC ....((((((((......((((((((..(((.(((((........)))))...))).....)))))))).....)))))))).. ( -32.90, z-score = -4.13, R) >droWil1.scaffold_180700 5427805 70 - 6630534 UUAAAGUU-UCCGUUAGCAACGCAGGAC------GCGUUCCUAGCAAUCAAAAUAUUAGCGCUUAACCGGAAAGCAC------- ......((-((((..((((((((.....------)))))....(((((......))).)))))....))))))....------- ( -15.20, z-score = -1.22, R) >consensus CUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGAUUAGUUACGGCCAACUGC ....((((((((............(((((...(((.....)))..)))))((((......))))..........)))))))).. (-12.39 = -14.22 + 1.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:35 2011