| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,701,223 – 3,701,343 |
| Length | 120 |
| Max. P | 0.774659 |

| Location | 3,701,223 – 3,701,343 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.51 |
| Shannon entropy | 0.63856 |
| G+C content | 0.56020 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -15.15 |
| Energy contribution | -14.35 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.94 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3701223 120 + 21146708 GGCGGCUUCAUUGCCUCCAGCUACGCCCUCUCACAUCCGGAGCGUGUGAAACACCUCAUACUAGCCGAUCCCUGGGGCUUCCCCGAGAAGCCCUCGGAUUCGACCAACGGCAAAACAAUA ((.(((......))).)).((((.((((..........)).))((((((......))))))))))(((..((.((((((((.....)))))))).))..))).................. ( -41.00, z-score = -2.60, R) >droSim1.chr2R 2428106 120 + 19596830 GGCGGCUUCAUUGCCUCCAGCUACGCCCUCUCACACCCGGAGCGUGUGAAACACCUCAUACUGGCCGAUCCCUGGGGCUUCCCCGAGAAGCCCUCGGACUCGACCAACGGCAAAACAAUA ((.(((......))).))......(((((((.......)))).((((((......))))))(((.(((..((.((((((((.....)))))))).))..))).)))..)))......... ( -44.10, z-score = -3.02, R) >droSec1.super_1 1357527 120 + 14215200 GGCGGCUUCAUUGCCUCCAGCUACGCCCUCUCACACCCGGAGCGUGUGAAACACCUCAUACUGGCCGAUCCCUGGGGCUUCCCCGAGAAGCCCUCGGACUCGACCAACGGCAAAACAAUA ((.(((......))).))......(((((((.......)))).((((((......))))))(((.(((..((.((((((((.....)))))))).))..))).)))..)))......... ( -44.10, z-score = -3.02, R) >droYak2.chr2L 16373419 120 + 22324452 GGCGGCUUCAUUGCCUCCAGCUACGCCCUUUCACACCCGGAGCGUGUGAAACACCUCAUACUGGCCGAUCCCUGGGGCUUCCCUGAGAAGCCCUCGGACUCGACCAACGGCAAAACAAUU ((.(((......))).))......(((.((((((((.......))))))))..........(((.(((..((.((((((((.....)))))))).))..))).)))..)))......... ( -45.20, z-score = -3.45, R) >droEre2.scaffold_4929 7733984 120 - 26641161 GGCGGCUUCAUUGCCUCCAGCUACGCCCUUUCACACCCGGAGCGUGUGAAACACCUCAUACUGGCCGAUCCCUGGGGCUUCCCCGAGAAGCCCUCGGACUCGACCAAUGGCAAAACAAUU ((.(((......))).))......(((.((((((((.......))))))))..........(((.(((..((.((((((((.....)))))))).))..))).)))..)))......... ( -44.90, z-score = -3.25, R) >droAna3.scaffold_13266 16046926 120 + 19884421 GGCGGUUUCAUCGCCUCUAGCUACGCCCUAUCAUAUCCAGAGCGUGUGCGACACUUAAUACUGGCCGAUCCGUGGGGCUUCCCCGAGAAGCCCUCCGACUCGACCAACACCAAACAGAUA ((((((...))))))(((.(((((((.((.........)).))))).))............(((.(((..((..(((((((.....)))))))..))..))).))).........))).. ( -39.40, z-score = -2.62, R) >dp4.chr3 17230076 120 - 19779522 GGUGGCUUCAUAGCCUCCAGCUACGCCUUGAGCUAUCCAGAGCGCGUCAAGCAUCUGGUGCUGGCUGACCCAUGGGGAUUCCCCGAGAAACCCACAGAUCCCAACAACACCAAACAGAUU (((((((....)))(((((((((((((..(((((......)))((.....)).)).)))).))))))......((((...)))))))....................))))......... ( -35.80, z-score = -0.86, R) >droPer1.super_2 6296069 120 - 9036312 GGUGGCUUCAUAGCCUCCAGCUACGCCUUGAGCUAUCCAGAGCGCGUCAAGCAUCUGGUGCUGGCUGACCCAUGGGGAUUCCCCGAGAAACCCACAGAUCCCAACAACACCAAACAGAUU (((((((....)))(((((((((((((..(((((......)))((.....)).)).)))).))))))......((((...)))))))....................))))......... ( -35.80, z-score = -0.86, R) >droWil1.scaffold_180745 1825101 120 + 2843958 GGCGGUUUCAUUGCAUCCAGCUAUGCGUUGAGCUAUCCGGAGCGAGUGAAACAUCUGAUAUUGGCUGAUCCUUGGGGUUUUCCAGAGAAACCAACGGAUUCAUCGAAUGGAAAACAGAUU ....((((((((((.(((((((........))))....))))).))))))))(((((.((((.(.((((((((((..((((...))))..)))).))).))).).)))).....))))). ( -33.50, z-score = -0.36, R) >droVir3.scaffold_12875 951082 120 + 20611582 GGCGGUUUCAUUGCCUCCAGCUAUGCACUUAGCUAUCCGGAGCGCGUUAGACACCUAAUACUGGCCGAUCCGUGGGGCUUUCCCGAGAAGCCCGCCGACUCGACGAACACCAAACAGAUU ...((((((...(((((((((((......)))))....)))).))(((((....)))))......(((..((..(((((((.....)))))))..))..)))..))).)))......... ( -35.40, z-score = -0.58, R) >droGri2.scaffold_15245 8308740 120 - 18325388 GGCGGCUUCAUUGCCUCCAGCUAUGCGUUGAGCUAUCCGGAGCGUGUACGCCAUCUGAUUCUAGCUGAUCCUUGGGGCUUUCCCGAGAAACCCACCGAUUCGACGAAUACCAAACAGAUC ((.(((......))).)).(.(((.((((((((((..(((((((....)))..))))....)))))((((..((((..(((....)))..))))..))))))))).))).)......... ( -33.70, z-score = -0.41, R) >droMoj3.scaffold_6496 3988628 120 + 26866924 GGCGGUUUCAUUGCGUCCAGCUAUGCCCUGAGCUAUCCGGAGCGUGUUCGACAUCUGAUACUGGCCGAUCCCUGGGGAUUUCCCGAGAAACCCGCCGAAUCGACGAAUACCAAACAGAUU (((((((((.....(((.(((.((((((.((....)).)).))))))).))).........(((..(((((....)))))..))).)))).)))))........................ ( -36.00, z-score = -0.53, R) >anoGam1.chrX 14273670 120 + 22145176 GGCGGCUUCAUUGCGGCGAGCUACGCGCUCUCCUACCCGGACCGGCUGCGCCAUCUCAUACUGGCCGACCCGUGGGGCAUGCCGGAGAAGCCGAAGGAGUUCGAAAACAAUGCGCGCAUC .((.((......)).))......(((((.(((((...(((.((((((((((((........))))...((....))))).))))).....))).)))))............))))).... ( -44.22, z-score = 0.72, R) >apiMel3.GroupUn 339185450 114 - 399230636 GGUGGUUUUCUUGCAGCAUCAUAUAGCAUGCAAUAUCCCGAAAGAAUUAAACACCUGAUUCUUGCCGAUCCAUGGGGUUUUCCGGAAAGACCUGUAGAUCGAAUUUCCAGAGUU------ (((((..(..(((((((........)).))))).)..))..((((((((......)))))))))))((((.(((((.((((....)))).))))).))))..............------ ( -26.90, z-score = -0.23, R) >triCas2.ChLG7 6243941 111 + 17478683 GGGGGUUUCCUUGCCGCUUCUUACGCUUUGAGUUACCCAGAUCAUGUCAAACAUUUGGUUUUGGCCGAUCCGUGGGGUUUCCCCGAGCGGCC-GCAG---GAAUUCAAAGAGUUG----- ((.(((......))).))....((.(((((((((.((.((((((((.....))..)))))).(((((.....(((((...)))))..)))))-...)---)))))))))).))..----- ( -35.20, z-score = -0.28, R) >consensus GGCGGCUUCAUUGCCUCCAGCUACGCCCUGACAUAUCCGGAGCGUGUGAAACACCUGAUACUGGCCGAUCCCUGGGGCUUCCCCGAGAAGCCCUCGGACUCGACCAACAGCAAACAGAUU ...((((((............................(((...(((((........)))))...)))......(((....)))...))))))............................ (-15.15 = -14.35 + -0.80)

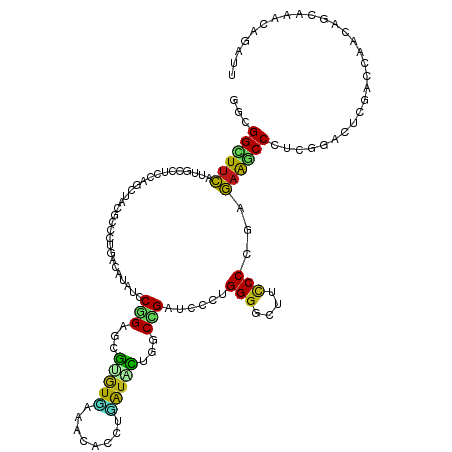

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:33 2011