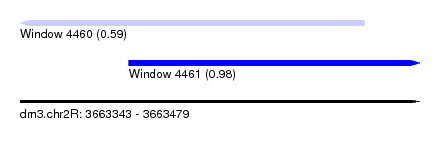
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,663,343 – 3,663,479 |
| Length | 136 |
| Max. P | 0.978839 |

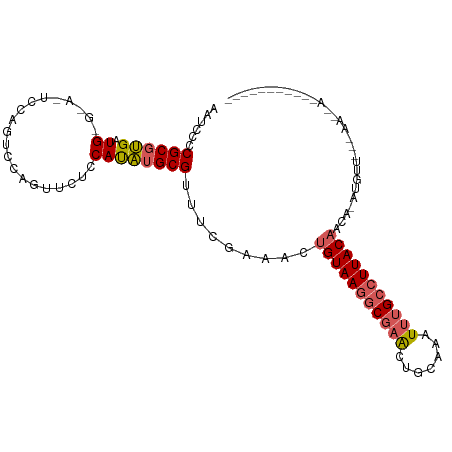
| Location | 3,663,343 – 3,663,460 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Shannon entropy | 0.31209 |
| G+C content | 0.45357 |
| Mean single sequence MFE | -32.69 |
| Consensus MFE | -18.57 |
| Energy contribution | -22.10 |
| Covariance contribution | 3.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3663343 117 - 21146708 GUAAGGCAAAUCUGCAGUUCGCCUUACAGUUUCGAAACGCAUAUGUAGAACUGGAUUGGACUCCUCAUCACGCGGGGAUUUCCACUCAGAUUCGAAUCUCGACAGAGAUUUGCAAAU (((((((.(((.....))).)))))))...........((.......(((((((..((((.(((((.......)))))..)))).)))).)))(((((((....))))))))).... ( -35.10, z-score = -2.44, R) >droSim1.chr2R 2397339 117 - 19596830 GUAAGGCAAAUUUGCAGUUCGCCUUACAGUUUCGAAACGCAUAUGCAGAAGUGGACUGGAGUUCCCAUCACGCGGGGAUUUCCACUCAGAUUCGAAUCUCGCCAGAGAUUUGCAAAU (((((((.(((.....))).)))))))................(((.(((...((.(((((.((((........)))).))))).))...)))(((((((....))))))))))... ( -38.50, z-score = -3.05, R) >droSec1.super_1 1318073 117 - 14215200 GUAAGGCAAAUUUGCAGUUCGCCUUACAGUUUCGAAACGCAUAUGCAGAACUGGACUGGAGUUCCCAUCACGCGGGGAUUUCCACUCAGAUUCGAAUCUCGCCAGAGAUUUGCAAAU (((((((.(((.....))).)))))))................(((.(((((((..(((((.((((........)))).))))).)))).)))(((((((....))))))))))... ( -38.70, z-score = -3.26, R) >droYak2.chr2L 16334284 99 - 22324452 GUAAGGCAAAUUUGCAGCUCGCCUUACAUUUUUGAAACGCAUGUGGAGAACUCAA--------CUCAUCACGCGGGGAUUUCCAGUCAGAUUCGA----------AGAUUUGCAAAU (((((((.............)))))))(((((((((.(((.((..(((.......--------)))..)).))).((....)).......)))))----------))))........ ( -24.32, z-score = -0.96, R) >droEre2.scaffold_4929 7695095 109 + 26641161 GUAAGGCUUAUUUGCAGUUCGAUUUACAUUUUCGAGACGCUCAUGGAGAACUCAG--------CUCACCGUGCGGGGAUUUCCAGUCAGAUUCGAAUCUCGCUAAAGAUUUGCAAAU .........((((((((.((..........((((((.(.....((((((.(((.(--------(.......)).))).))))))....).))))))..........)).)))))))) ( -26.85, z-score = -0.39, R) >consensus GUAAGGCAAAUUUGCAGUUCGCCUUACAGUUUCGAAACGCAUAUGCAGAACUGGA_UGGA_U_CUCAUCACGCGGGGAUUUCCACUCAGAUUCGAAUCUCGCCAGAGAUUUGCAAAU (((((((.(((.....))).)))))))...........((.......(((((((...(((.(((((.......))))).)))...)))).)))(((((((....))))))))).... (-18.57 = -22.10 + 3.53)

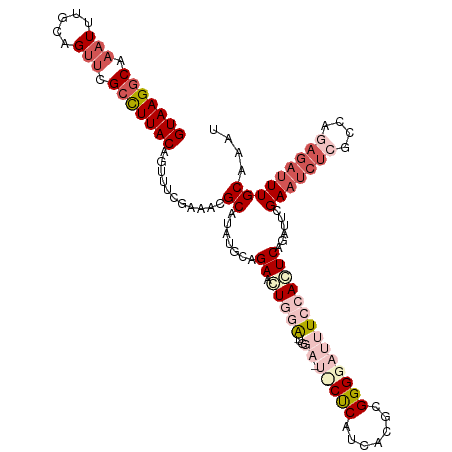
| Location | 3,663,380 – 3,663,479 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.63 |
| Shannon entropy | 0.52433 |
| G+C content | 0.43939 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -13.47 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3663380 99 + 21146708 AAUCCCCGCGUGAUGAGGAGUCCAAUCCAGUUCUACAUAUGCGUUUCGAAACUGUAAGGCGAACUGCAGAUUUGCCUUACAACAAAUGUUCGCAACGAA---------- .......(((((.((((((.(.......).)))).)))))))(((.((((..((((((((((((....).))))))))))).......)))).)))...---------- ( -24.90, z-score = -1.43, R) >droSim1.chr2R 2397376 84 + 19596830 AAUCCCCGCGUGAUGGGAACUCCAGUCCACUUCUGCAUAUGCGUUUCGAAACUGUAAGGCGAACUGCAAAUUUGCCUUACAACA------------------------- ......((((((.((((((...........)))).)))))))).........(((((((((((.......)))))))))))...------------------------- ( -20.40, z-score = -1.25, R) >droSec1.super_1 1318110 84 + 14215200 AAUCCCCGCGUGAUGGGAACUCCAGUCCAGUUCUGCAUAUGCGUUUCGAAACUGUAAGGCGAACUGCAAAUUUGCCUUACAACA------------------------- ......((((((.((((((((.......)))))).)))))))).........(((((((((((.......)))))))))))...------------------------- ( -25.30, z-score = -2.56, R) >droYak2.chr2L 16334311 100 + 22324452 AAUCCCCGCGUGAUG--------AGUUGAGUUCUCCACAUGCGUUUCAAAAAUGUAAGGCGAGCUGCAAAUUUGCCUUACAA-ACAUGUUCCAAAUGAAUAUGUAAAUA ......((((((..(--------((.......)))..)))))).........(((((((((((.......))))))))))).-((((((((.....))))))))..... ( -29.90, z-score = -4.48, R) >droEre2.scaffold_4929 7695132 100 - 26641161 AAUCCCCGCACGGUG--------AGCUGAGUUCUCCAUGAGCGUCUCGAAAAUGUAAAUCGAACUGCAAAUAAGCCUUACUA-ACAUGUUAAAAAUAAGUAUGUACGUA .........((((((--------((((..((((.....))))((.((((.........))))))........)).)))))..-((((((((....))).))))).))). ( -14.80, z-score = 0.46, R) >consensus AAUCCCCGCGUGAUG_G_A_UCCAGUCCAGUUCUCCAUAUGCGUUUCGAAACUGUAAGGCGAACUGCAAAUUUGCCUUACAACA_AUGUU___AA__A___________ ......((((((.((....................)))))))).........(((((((((((.......)))))))))))............................ (-13.47 = -14.03 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:30 2011