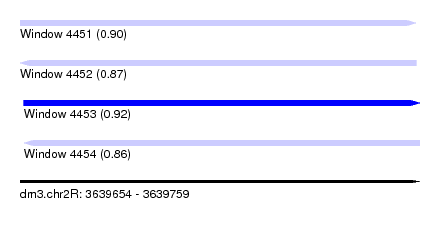
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,639,654 – 3,639,759 |
| Length | 105 |
| Max. P | 0.923176 |

| Location | 3,639,654 – 3,639,758 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.24 |
| Shannon entropy | 0.15229 |
| G+C content | 0.50233 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -27.36 |
| Energy contribution | -26.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
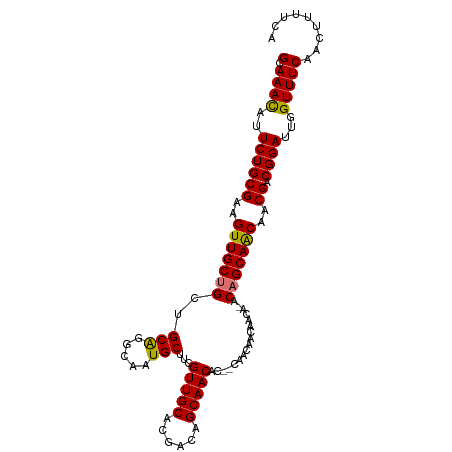
>dm3.chr2R 3639654 104 + 21146708 GCAAACAUUCUGCGAAGUUGCUGCUGCGGGCAAUGCUUCGUUGCACGACAGCAACACAGCAACACCAACA-ACAGCAGCGACGACGGAUUGGUUUCAACUUUUCG (.((((..((((((..(((((((.((..((...((((..(((((......)))))..))))...))..))-.)))))))..)).))))...)))))......... ( -36.20, z-score = -2.94, R) >droSec1.super_1 1295125 101 + 14215200 GCAAACAUUCUGCGAAGUUGCUGCUGCAGGCAAUGCUUCGUUGCACGACAGCAACAC--CAGCAACAACA--CAGCAGCGACGACGGAUUGGUUUCAACUUUUCG (.((((..((((....((((((((((.......((((..(((((......)))))..--.))))......--))))))))))..))))...)))))......... ( -32.52, z-score = -1.95, R) >droYak2.chr2L 16311683 105 + 22324452 GCAAAUAUUCUGCGAAGUUGCGGCUGCGGGCAAUGCUUCGUUGCACGACUGCAACACAGCAACACAGCCAUACAGCAACAACGACGGAUUGGUUUCAACUUUUCA (.((((..((((((..((((((((((.(.....((((..((((((....))))))..)))).).))))).....)))))..)).))))...)))))......... ( -31.80, z-score = -1.82, R) >droEre2.scaffold_4929 7672354 100 - 26641161 GCAAACAUUCUGCGAAGUUGCUGCUGCAGGCAAUGCUUCGUUGCACGACAGCAACAC---AGCAACA-CA-ACAGCAACAACGACGGAUUGGUUUCAACUUUUCA (.((((..((((((..(((((((.((....)).((((..(((((......)))))..---))))...-..-.)))))))..)).))))...)))))......... ( -29.40, z-score = -1.75, R) >consensus GCAAACAUUCUGCGAAGUUGCUGCUGCAGGCAAUGCUUCGUUGCACGACAGCAACAC__CAACAACAACA_ACAGCAACAACGACGGAUUGGUUUCAACUUUUCA (.((((..((((((..(((((((..(((.....)))...(((((......))))).................)))))))..)).))))...)))))......... (-27.36 = -26.93 + -0.44)

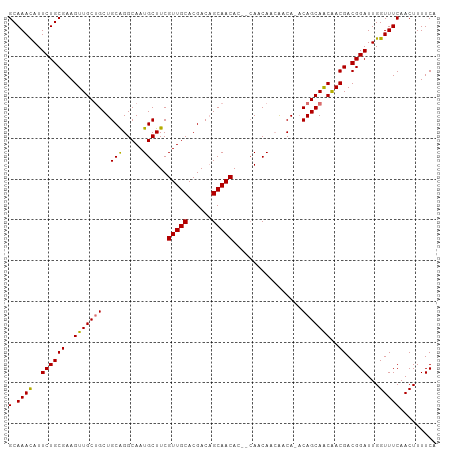
| Location | 3,639,654 – 3,639,758 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Shannon entropy | 0.15229 |
| G+C content | 0.50233 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -30.59 |
| Energy contribution | -31.65 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
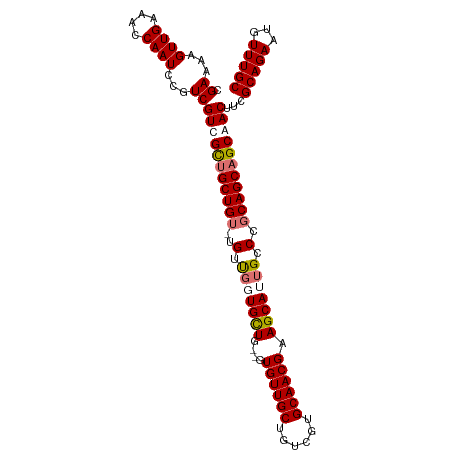
>dm3.chr2R 3639654 104 - 21146708 CGAAAAGUUGAAACCAAUCCGUCGUCGCUGCUGU-UGUUGGUGUUGCUGUGUUGCUGUCGUGCAACGAAGCAUUGCCCGCAGCAGCAACUUCGCAGAAUGUUUGC (((...((((....))))...)))....((((((-(((.(((..((((.((((((......)))))).))))..))).))))))))).....(((((...))))) ( -38.00, z-score = -2.09, R) >droSec1.super_1 1295125 101 - 14215200 CGAAAAGUUGAAACCAAUCCGUCGUCGCUGCUG--UGUUGUUGCUG--GUGUUGCUGUCGUGCAACGAAGCAUUGCCUGCAGCAGCAACUUCGCAGAAUGUUUGC (((...((((....))))...)))...((((..--(((((((((.(--((..((((.(((.....)))))))..))).))))))))).....))))......... ( -34.20, z-score = -1.13, R) >droYak2.chr2L 16311683 105 - 22324452 UGAAAAGUUGAAACCAAUCCGUCGUUGUUGCUGUAUGGCUGUGUUGCUGUGUUGCAGUCGUGCAACGAAGCAUUGCCCGCAGCCGCAACUUCGCAGAAUAUUUGC .((...((((....))))...))(((((.(((((..(((.(((((....(((((((....))))))).))))).))).))))).)))))...(((((...))))) ( -35.20, z-score = -1.84, R) >droEre2.scaffold_4929 7672354 100 + 26641161 UGAAAAGUUGAAACCAAUCCGUCGUUGUUGCUGU-UG-UGUUGCU---GUGUUGCUGUCGUGCAACGAAGCAUUGCCUGCAGCAGCAACUUCGCAGAAUGUUUGC .((...((((....))))...))(((((((((((-.(-(..((((---.((((((......)))))).))))..))..)))))))))))...(((((...))))) ( -36.60, z-score = -2.85, R) >consensus CGAAAAGUUGAAACCAAUCCGUCGUCGCUGCUGU_UGUUGGUGCUG__GUGUUGCUGUCGUGCAACGAAGCAUUGCCCGCAGCAGCAACUUCGCAGAAUGUUUGC .((...((((....))))...))(((((((((((....(((((((....((((((......)))))).)))))))...)))))))))))...(((((...))))) (-30.59 = -31.65 + 1.06)

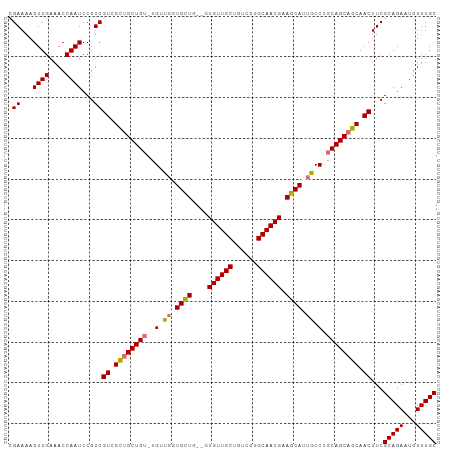
| Location | 3,639,655 – 3,639,759 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.24 |
| Shannon entropy | 0.15229 |
| G+C content | 0.49257 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -26.96 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3639655 104 + 21146708 CAAACAUUCUGCGAAGUUGCUGCUGCGGGCAAUGCUUCGUUGCACGACAGCAACACAGCAACACCAACA-ACAGCAGCGACGACGGAUUGGUUUCAACUUUUCGA ....((.((((((..(((((((.((..((...((((..(((((......)))))..))))...))..))-.)))))))..)).)))).))............... ( -35.80, z-score = -2.92, R) >droSec1.super_1 1295126 101 + 14215200 CAAACAUUCUGCGAAGUUGCUGCUGCAGGCAAUGCUUCGUUGCACGACAGCAACAC--CAGCAACAACA--CAGCAGCGACGACGGAUUGGUUUCAACUUUUCGA ....((.((((....((((((((((.......((((..(((((......)))))..--.))))......--))))))))))..)))).))............... ( -32.12, z-score = -1.91, R) >droYak2.chr2L 16311684 105 + 22324452 CAAAUAUUCUGCGAAGUUGCGGCUGCGGGCAAUGCUUCGUUGCACGACUGCAACACAGCAACACAGCCAUACAGCAACAACGACGGAUUGGUUUCAACUUUUCAA ...........((..((((..((((..(((..((((..((((((....))))))..)))).....)))...))))..))))..))((..(((....)))..)).. ( -31.50, z-score = -2.07, R) >droEre2.scaffold_4929 7672355 100 - 26641161 CAAACAUUCUGCGAAGUUGCUGCUGCAGGCAAUGCUUCGUUGCACGACAGCAACAC---AGCAACA-CA-ACAGCAACAACGACGGAUUGGUUUCAACUUUUCAA ....((.((((((..(((((((.((....)).((((..(((((......)))))..---))))...-..-.)))))))..)).)))).))............... ( -29.00, z-score = -1.99, R) >consensus CAAACAUUCUGCGAAGUUGCUGCUGCAGGCAAUGCUUCGUUGCACGACAGCAACAC__CAACAACAACA_ACAGCAACAACGACGGAUUGGUUUCAACUUUUCAA .((((..((((((..(((((((..(((.....)))...(((((......))))).................)))))))..)).))))...))))........... (-26.96 = -26.52 + -0.44)

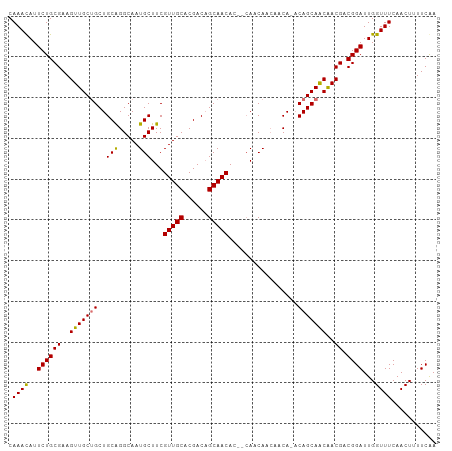
| Location | 3,639,655 – 3,639,759 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Shannon entropy | 0.15229 |
| G+C content | 0.49257 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -29.09 |
| Energy contribution | -30.15 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
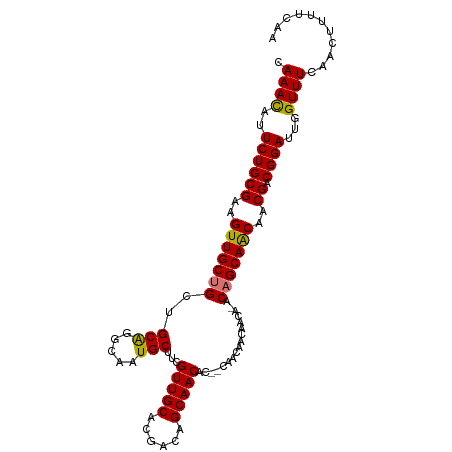
>dm3.chr2R 3639655 104 - 21146708 UCGAAAAGUUGAAACCAAUCCGUCGUCGCUGCUGU-UGUUGGUGUUGCUGUGUUGCUGUCGUGCAACGAAGCAUUGCCCGCAGCAGCAACUUCGCAGAAUGUUUG .(((...((((....))))...)))..((((((((-(((.(((..((((.((((((......)))))).))))..))).))))))))).....)).......... ( -37.10, z-score = -2.23, R) >droSec1.super_1 1295126 101 - 14215200 UCGAAAAGUUGAAACCAAUCCGUCGUCGCUGCUG--UGUUGUUGCUG--GUGUUGCUGUCGUGCAACGAAGCAUUGCCUGCAGCAGCAACUUCGCAGAAUGUUUG .(((...((((....))))...)))...((((..--(((((((((.(--((..((((.(((.....)))))))..))).))))))))).....))))........ ( -34.30, z-score = -1.59, R) >droYak2.chr2L 16311684 105 - 22324452 UUGAAAAGUUGAAACCAAUCCGUCGUUGUUGCUGUAUGGCUGUGUUGCUGUGUUGCAGUCGUGCAACGAAGCAUUGCCCGCAGCCGCAACUUCGCAGAAUAUUUG ..((...((((....))))...))(((((.(((((..(((.(((((....(((((((....))))))).))))).))).))))).)))))............... ( -33.70, z-score = -1.68, R) >droEre2.scaffold_4929 7672355 100 + 26641161 UUGAAAAGUUGAAACCAAUCCGUCGUUGUUGCUGU-UG-UGUUGCU---GUGUUGCUGUCGUGCAACGAAGCAUUGCCUGCAGCAGCAACUUCGCAGAAUGUUUG ..((...((((....))))...))(((((((((((-.(-(..((((---.((((((......)))))).))))..))..)))))))))))............... ( -35.10, z-score = -2.69, R) >consensus UCGAAAAGUUGAAACCAAUCCGUCGUCGCUGCUGU_UGUUGGUGCUG__GUGUUGCUGUCGUGCAACGAAGCAUUGCCCGCAGCAGCAACUUCGCAGAAUGUUUG ..((...((((....))))...))(((((((((((....(((((((....((((((......)))))).)))))))...)))))))))))............... (-29.09 = -30.15 + 1.06)


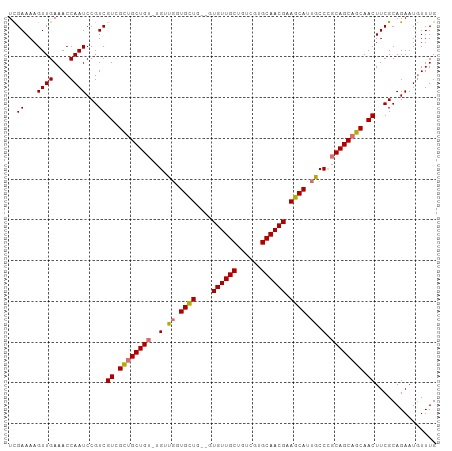
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:25 2011