| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,618,752 – 3,618,850 |
| Length | 98 |
| Max. P | 0.772535 |

| Location | 3,618,752 – 3,618,850 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.81 |
| Shannon entropy | 0.30397 |
| G+C content | 0.49783 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -17.44 |
| Energy contribution | -19.72 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
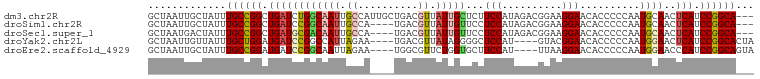
>dm3.chr2R 3618752 98 + 21146708 ---UGCCGGAUGAGUUGCAUUGGGGGUGUUCCUUCCGUCUAUGGAAGAGCAAUAACGUCAGCAAUGGCAAUUGCCAGAUCAGCCGGCAAAUAGCAAUUAGC ---((((((.((((((((..(((.(.(((((.(((((....))))))))))....).))))))))(((....)))...))).))))))............. ( -35.20, z-score = -2.36, R) >droSim1.chr2R 2347973 94 + 19596830 ---UGCCGGAUGAGUUGCAUUGGGGGUGUUCCUUCCGUCUAUGGAGGAACAAUAACGUCA----UGGCAAUUGCCGGAUCAGCCGGCAAAUAGCAAUUAGC ---((((((.(((.......(((.(.(((((((.(((....))))))))))....).)))----((((....))))..))).))))))............. ( -33.20, z-score = -1.91, R) >droSec1.super_1 1274207 94 + 14215200 ---UGCCGGAUGAGUUGCAUUGGGGGUGUUCCUUCCGUCUAUGGAGGAACAAUAACGUCA----UGGCAAUUGUCGCAUCAGCCGGCAAAUAGUCAUUAGC ---((((((.(((..(((..(((.(.(((((((.(((....))))))))))....).)))----.(((....))))))))).))))))............. ( -33.30, z-score = -2.45, R) >droYak2.chr2L 16291400 93 + 22324452 UAGUGCCGGAUGAGUUCCAUUGGGGGUGUUCCGUAC----AUGGAGCCCCUAUAACGUCA----UUCUAAUGGCCGGAUCAUCCAGCAAAUAACAAUUAGC ...(((.((((((..(((..(((((((..((((...----.)))))))))))....((((----(....))))).))))))))).)))............. ( -33.00, z-score = -2.84, R) >droEre2.scaffold_4929 7652039 93 - 26641161 UACUGCCGGAUGGGUUCCAUUGGGGGUGUUCCUUAA----AUGGAAGCACCAGAACGCCA----UUCUAAUUGCCGGAUCAUCCGGCAAAUAGCAAUUAGC ...(((.((((((((((.......((((((((....----..)).)))))).)))).)))----)))...(((((((.....)))))))...)))...... ( -35.60, z-score = -3.05, R) >consensus ___UGCCGGAUGAGUUGCAUUGGGGGUGUUCCUUCCGUCUAUGGAGGAACAAUAACGUCA____UGGCAAUUGCCGGAUCAGCCGGCAAAUAGCAAUUAGC ...((((((.(((.......(((.(.(((((((((.......)))))))))....).)))....((((....))))..))).))))))............. (-17.44 = -19.72 + 2.28)



| Location | 3,618,752 – 3,618,850 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Shannon entropy | 0.30397 |
| G+C content | 0.49783 |
| Mean single sequence MFE | -30.29 |
| Consensus MFE | -13.69 |
| Energy contribution | -15.01 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3618752 98 - 21146708 GCUAAUUGCUAUUUGCCGGCUGAUCUGGCAAUUGCCAUUGCUGACGUUAUUGCUCUUCCAUAGACGGAAGGAACACCCCCAAUGCAACUCAUCCGGCA--- ((.....))....((((((.(((..((((....))))((((............((((((......))))))............)))).))).))))))--- ( -28.95, z-score = -1.85, R) >droSim1.chr2R 2347973 94 - 19596830 GCUAAUUGCUAUUUGCCGGCUGAUCCGGCAAUUGCCA----UGACGUUAUUGUUCCUCCAUAGACGGAAGGAACACCCCCAAUGCAACUCAUCCGGCA--- ((.....))....((((((.(((...(((....))).----((.((((..(((((((((......)).))))))).....))))))..))).))))))--- ( -31.00, z-score = -3.00, R) >droSec1.super_1 1274207 94 - 14215200 GCUAAUGACUAUUUGCCGGCUGAUGCGACAAUUGCCA----UGACGUUAUUGUUCCUCCAUAGACGGAAGGAACACCCCCAAUGCAACUCAUCCGGCA--- .............((((((.(((((((((........----....)))..(((((((((......)).)))))))........)))..))).))))))--- ( -26.40, z-score = -2.24, R) >droYak2.chr2L 16291400 93 - 22324452 GCUAAUUGUUAUUUGCUGGAUGAUCCGGCCAUUAGAA----UGACGUUAUAGGGGCUCCAU----GUACGGAACACCCCCAAUGGAACUCAUCCGGCACUA .............((((((((((((((..(((....)----))........((((.(((..----....)))....))))..))))..))))))))))... ( -28.70, z-score = -1.64, R) >droEre2.scaffold_4929 7652039 93 + 26641161 GCUAAUUGCUAUUUGCCGGAUGAUCCGGCAAUUAGAA----UGGCGUUCUGGUGCUUCCAU----UUAAGGAACACCCCCAAUGGAACCCAUCCGGCAGUA ....((((((..(((((((.....)))))))...(.(----(((.(((((((((.((((..----....))))))))......))))))))).))))))). ( -36.40, z-score = -3.75, R) >consensus GCUAAUUGCUAUUUGCCGGCUGAUCCGGCAAUUGCCA____UGACGUUAUUGUUCCUCCAUAGACGGAAGGAACACCCCCAAUGCAACUCAUCCGGCA___ .............((((((.(((((((((................)))...((((((((......))).))))).........)))..))).))))))... (-13.69 = -15.01 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:18 2011