| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,617,370 – 3,617,463 |
| Length | 93 |
| Max. P | 0.859036 |

| Location | 3,617,370 – 3,617,463 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 54.85 |
| Shannon entropy | 0.92142 |
| G+C content | 0.53383 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.08 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3617370 93 - 21146708 ----UGUCGCUCGGCACCAU-UGGUGAUUCAUU--CGCGGAGUU-----------CUGGGCUCUGUGAACACCAAACCCACGCACACAAGUCGAAACUGGCUAAAGCCGGA ----......(((((....(-(((((.....((--((((((((.-----------....))))))))))))))))......(....)..)))))..(((((....))))). ( -31.30, z-score = -1.51, R) >droAna3.scaffold_13266 10124027 83 - 19884421 ----------------------UGUCUGCUGGCUUUGCAGAGUUCUCAACAGUUAG------UGAUAACUCAUACCCACCUGUGUUCAGGUUGAAACUGGCUCUGGCCAGA ----------------------......((((((..((..((((.((((((((((.------...))))).........(((....)))))))))))).))...)))))). ( -26.60, z-score = -1.26, R) >droEre2.scaffold_4929 7650576 104 + 26641161 ----UGUUGCUCGGCACAAU-UGGUGAAUCAUC--CGCGGAGUUCUGGGCGCCCAGAUAGCUUUAUAAGCACCAUACCCACGCACACAAGAUGGAACUGGCUAAAGCCGGA ----((((((..((......-(((((.......--.((.....(((((....)))))..))........)))))...))..))).)))........(((((....))))). ( -28.59, z-score = 0.26, R) >droYak2.chr2L 16290034 111 - 22324452 UGUCGCUCGGUACCAUUGGUGAUUCAUCCAGAUUCAGCGGAGUUCUGGGCAACCAGAUAGCUUUAUAAUCACCAUACCCACGCACACAAGAUGGAACUGGCUAAAGCCGGA (((.((..((((....((((((((..(((.........)))..(((((....))))).........))))))))))))...)))))..........(((((....))))). ( -34.00, z-score = -1.82, R) >droSec1.super_1 1272830 110 - 14215200 -UGUCGCUCCGCACCAUUGGUGGGUGAUUCAUCGGAGCGGAGUUCUGGGCACCCAGAUAGCUUUGUGAACACCAAACCCACGCACACAAGCCGAAACUGGCUAAAGCCGGA -(((.((....((((...))))((((.((((.((((((.....(((((....)))))..))))))))))))))........)))))..........(((((....))))). ( -39.00, z-score = -1.39, R) >droSim1.chr2R 2346655 110 - 19596830 -UGUCGCUCCGCACCAUUGGUGGGUGAUUCAUCGGAGCGGAGUUCUGGGCACCCAGAUAGCUUUGUGAACACCAAACCCACGCACACAAGUCGAAACUGGCUAAAGCCGGA -(((.((....((((...))))((((.((((.((((((.....(((((....)))))..))))))))))))))........)))))..........(((((....))))). ( -39.00, z-score = -1.48, R) >droPer1.super_2 6208512 92 + 9036312 --------------UAUCAG-ACGUGACUCACU--CGCGGAGUUGGAGAUAACUUUUGGGCAC--UCAGCCACACACAAGAUUCCGCAGGCCGAAACUGGCUUUGGCCGGC --------------.....(-(.(((...))))--)((((((((((((....))))(((((..--...))).)).....)))))))).(((((((......)))))))... ( -28.10, z-score = -1.12, R) >dp4.chr3 17138082 92 + 19779522 --------------UAUCAG-ACGUGACUCACU--CGCGGAGUUGGAGAUAACUUUUGGGCAC--UCAGCCACACACAAGAUUCCACAGGCCGAAACUGGCUUUGGCCGGC --------------......-.(((((.....)--))))..((.((((..........(((..--...)))..........)))))).(((((((......)))))))... ( -24.15, z-score = -0.30, R) >consensus _____G___C_C__CAUCAG_AGGUGACUCAUC__CGCGGAGUUCUGGGCAACCAGAUAGCUUUAUAAACACCAAACCCACGCACACAAGCCGAAACUGGCUAAAGCCGGA ....................................((......((((....))))...))...................................(((((....))))). (-10.65 = -10.08 + -0.58)

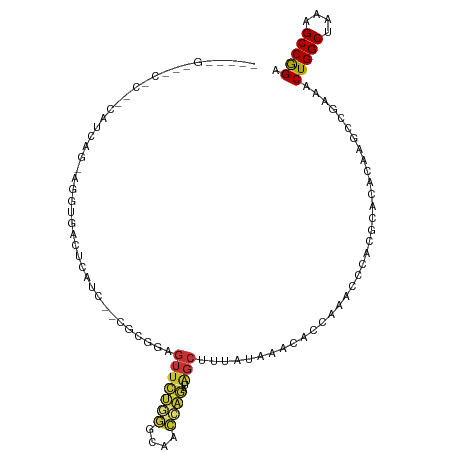

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:16 2011