
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,292,518 – 2,292,611 |
| Length | 93 |
| Max. P | 0.955988 |

| Location | 2,292,518 – 2,292,611 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.29 |
| Shannon entropy | 0.51132 |
| G+C content | 0.44952 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -9.62 |
| Energy contribution | -10.70 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2292518 93 + 23011544 -----AGCCAGGAC-AAGUUAAUGAGAAUCGUAUUUCAUUUGCAAUUAGCAA--GGAGCACACGCAGCCAUUACAUUA--CAUUGUC-CUGGCCUCCGGUAUCC---------- -----.((((((((-((..(((((.(((......)))..((((.....))))--((.((....))..))....)))))--..)))))-)))))...........---------- ( -28.70, z-score = -3.82, R) >droSim1.chr2L 2252872 94 + 22036055 -----AGCCAGGAC-AAGUUAAUGAGCAUCGUAUUUCAUUUGCAAUUAGCAAA-GGAGCACACGCAGCCAUUACAAUA--CAUUGUC-CUGGCCUCCGGUUUCC---------- -----.((((((((-(((((((((.((..(((..(((.(((((.....)))))-.)))...)))..)))))))....)--).)))))-)))))...........---------- ( -31.20, z-score = -4.52, R) >droEre2.scaffold_4929 2329550 75 + 26641161 -----GGCCAGGAC-AAGUUAAUGAGAAUCGUAUUUCAUUUGCAAUUAGCAAGAGGAGCACACGCA-----CGCAUCA--AAAUGUC-C------------------------- -----.....((((-(..((.(((.....(((.((((.(((((.....))))).))))...)))..-----..))).)--)..))))-)------------------------- ( -15.00, z-score = -1.02, R) >droYak2.chr2L 2274033 87 + 22324452 -----AGCCAGGAC-CAGUUAAUGAGAAUCGUAUUUCAUUUGCAAUUAGCAA--GGAGCACACGCA-----CGCAUCA--AAAUGUC-CUGGUCUCCUGCUCC----------- -----.((((((((-...((.(((.....(((..(((.(((((.....))))--))))...)))..-----..))).)--)...)))-)))))..........----------- ( -21.90, z-score = -1.30, R) >droAna3.scaffold_12916 10012343 110 - 16180835 UGCCAGGCCGGUUCUCAGUUAAUGAGAAUCUGAUUUAAUUUCCAAUUAGCAA--GGAACAC-CGCAAACA-CAUGAUAAUUAUUGUAACCAGUCGACAUUGUCCUGCCUGCUCC .((.((((.((((((..((((((..(((...........)))..))))))..--)))))..-........-...((((((.((((....))))....))))))).))))))... ( -21.70, z-score = -0.25, R) >consensus _____AGCCAGGAC_AAGUUAAUGAGAAUCGUAUUUCAUUUGCAAUUAGCAA__GGAGCACACGCA__CA_CACAUUA__AAUUGUC_CUGGCCUCCGGUUUCC__________ ......((((((((...((.(((((((......))))))).)).....((.............))...................))).)))))..................... ( -9.62 = -10.70 + 1.08)

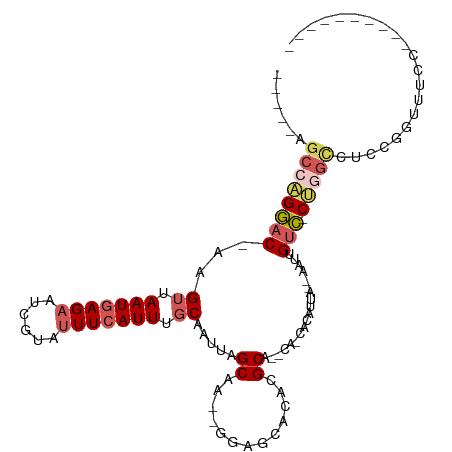
| Location | 2,292,518 – 2,292,611 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.29 |
| Shannon entropy | 0.51132 |
| G+C content | 0.44952 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -9.50 |
| Energy contribution | -10.78 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2292518 93 - 23011544 ----------GGAUACCGGAGGCCAG-GACAAUG--UAAUGUAAUGGCUGCGUGUGCUCC--UUGCUAAUUGCAAAUGAAAUACGAUUCUCAUUAACUU-GUCCUGGCU----- ----------..........((((((-(((((..--(((((....((...(((((..((.--((((.....))))..)).)))))..)).)))))..))-)))))))))----- ( -31.70, z-score = -3.48, R) >droSim1.chr2L 2252872 94 - 22036055 ----------GGAAACCGGAGGCCAG-GACAAUG--UAUUGUAAUGGCUGCGUGUGCUCC-UUUGCUAAUUGCAAAUGAAAUACGAUGCUCAUUAACUU-GUCCUGGCU----- ----------(....)....((((((-(((((.(--(....(((((((..(((((..((.-(((((.....))))).)).)))))..)).)))))))))-)))))))))----- ( -37.00, z-score = -4.97, R) >droEre2.scaffold_4929 2329550 75 - 26641161 -------------------------G-GACAUUU--UGAUGCG-----UGCGUGUGCUCCUCUUGCUAAUUGCAAAUGAAAUACGAUUCUCAUUAACUU-GUCCUGGCC----- -------------------------(-((((..(--(((((.(-----..(((((..((...((((.....))))..)).)))))...).))))))..)-)))).....----- ( -22.80, z-score = -4.01, R) >droYak2.chr2L 2274033 87 - 22324452 -----------GGAGCAGGAGACCAG-GACAUUU--UGAUGCG-----UGCGUGUGCUCC--UUGCUAAUUGCAAAUGAAAUACGAUUCUCAUUAACUG-GUCCUGGCU----- -----------........((.((((-(((...(--(((((.(-----..(((((..((.--((((.....))))..)).)))))...).))))))...-)))))))))----- ( -28.40, z-score = -2.30, R) >droAna3.scaffold_12916 10012343 110 + 16180835 GGAGCAGGCAGGACAAUGUCGACUGGUUACAAUAAUUAUCAUG-UGUUUGCG-GUGUUCC--UUGCUAAUUGGAAAUUAAAUCAGAUUCUCAUUAACUGAGAACCGGCCUGGCA ....(((((.((..........((((((....(((((.(((..-.(((.(((-(......--)))).)))))).))))))))))).((((((.....)))))))).)))))... ( -25.50, z-score = 0.25, R) >consensus __________GGAAACCGGAGACCAG_GACAAUG__UAAUGUG_UG__UGCGUGUGCUCC__UUGCUAAUUGCAAAUGAAAUACGAUUCUCAUUAACUU_GUCCUGGCU_____ ......................((((.(((......(((((.........(((((.......((((.....)))).....))))).....))))).....)))))))....... ( -9.50 = -10.78 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:47 2011