
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,612,387 – 3,612,477 |
| Length | 90 |
| Max. P | 0.806863 |

| Location | 3,612,387 – 3,612,477 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.98 |
| Shannon entropy | 0.70445 |
| G+C content | 0.48134 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -11.78 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3612387 90 - 21146708 GGCCAAGUGCUUUGUGACCUU----CUUCA-----UUUGUGACGCC---AACUGUGACGUAGGAAUAUUGAGCUGAGUGGCGUCAUAGUUUGCGUGAAAUCU ........((...((((....----..)))-----)..)).((((.---(((((((((((.((.........)).....))))))))))).))))....... ( -22.50, z-score = -0.06, R) >droSim1.chr2R 2340665 90 - 19596830 GGCCAAGUGUUCGGUGACCUU----CUUCA-----UUUGUGACGCC---AACUGUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCU .(((........)))......----.((((-----(((((((((((---(.((((...(((....)))...))))..))))))))))).....))))).... ( -25.60, z-score = -0.76, R) >droSec1.super_1 1267828 90 - 14215200 GGCCAAGUGUUCGGUGACCUU----CUUCA-----UUUGUGACGCC---AACUAUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCU ...((((((...(....)...----...))-----))))(.((((.---(((((((((((...................))))))))))).)))).)..... ( -25.01, z-score = -0.83, R) >droYak2.chr2L 16284959 87 - 22324452 GGCCAAGUGUUUUGUGACCUU----UUUC--------UGUGACGCC---AACUGUGACGUAGAAACAUUAAGCACUGUGGCGUCAUAGUUUGCGUGAAAUUU ((..(((.(((....))))))----..))--------..(.((((.---(((((((((((....(((........))).))))))))))).)))).)..... ( -24.90, z-score = -1.52, R) >droEre2.scaffold_4929 7645679 88 + 26641161 GGCCAAGUGUUUUGUGACCUU----CUUC-------UUGUGACGCC---AACUGUGACGUAGGAAUAUUAAGCACUGUGGCGUCAUAGUUUGCGUGAAAUCU ((..(((.(((....))))))----..))-------...(.((((.---(((((((((((.((...........))...))))))))))).)))).)..... ( -23.10, z-score = -0.78, R) >droAna3.scaffold_13266 10120032 102 - 19884421 GAGCAAGUGUUUUAUUAUUUUAAGGGUUCACCAAGUCCUAGACGUCGUGACCUAUGACGUAUGAAUGGAGAACCUGGUGGCGUCACAACUUGCAUGACGCCC ((((....))))...........((((((.(((..((....((((((((...))))))))..)).))).))))))...(((((((.........))))))). ( -30.40, z-score = -1.75, R) >droPer1.super_2 6204769 81 + 9036312 -GCCAAGUGCCCGG-----UU----UUCCA-----UUUGUGACGAC---AGCAGUGACGAUGG---AUUGACCAUGUCGGCGUCACUGUUUGCAUGACAUCU -(((....((..((-----((----.((((-----((.((.((...---....)).)))))))---)..))))..)).)))((((.((....)))))).... ( -22.90, z-score = -0.16, R) >droVir3.scaffold_12875 832095 93 - 20611582 AACAAAAUCCCUUAUCGCAGCGGCACUUUCUUGGGCCUCUGCCAGG---CCACGUGACGUCAUUGUCCUGCGGCUGUCGACGUCAUCGUUAGCAUG------ ................((((.(((.((.....))))).))))..((---((.((.((((....)))).)).))))((..(((....)))..))...------ ( -28.90, z-score = -1.30, R) >consensus GGCCAAGUGUUUUGUGACCUU____CUUCA_____UUUGUGACGCC___AACUGUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCU ..........................................(((....(((((((((((...................)))))))))))...)))...... (-11.78 = -12.22 + 0.44)

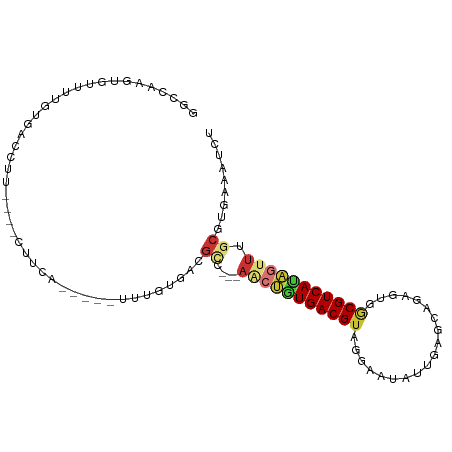
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:16 2011