
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,608,927 – 3,609,028 |
| Length | 101 |
| Max. P | 0.856852 |

| Location | 3,608,927 – 3,609,028 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 71.49 |
| Shannon entropy | 0.51633 |
| G+C content | 0.50878 |
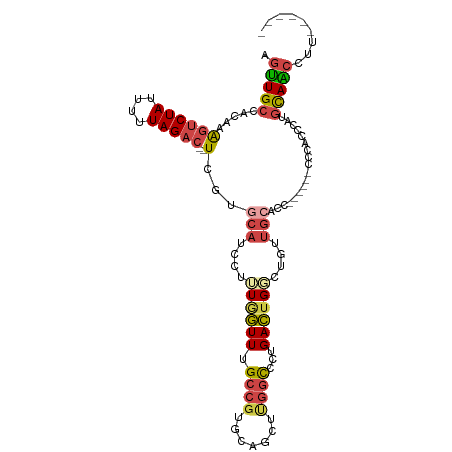
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -11.39 |
| Energy contribution | -12.37 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856852 |
| Prediction | RNA |
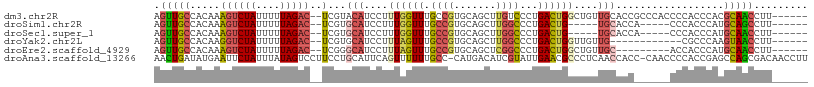
Download alignment: ClustalW | MAF
>dm3.chr2R 3608927 101 - 21146708 AGUUGCCACAAAGUCUAUUUUUAGAC--UCGUACAUCCUUUGGUUUGCCGUGCAGCUUGUCCCUGACUGGCUGUUGCACCGCCCACCCCACCCACGCAACCUU------ .(((((.....((((((....)))))--)............(((..((.(((((((..(((.......))).))))))).))..)))........)))))...------ ( -29.40, z-score = -3.30, R) >droSim1.chr2R 2336387 91 - 19596830 AGUUGCCACAAAGUCUAUUUUUAGAC--UCGUGCAUCCUUUGGUUUGCCGUGCAGCUUGGCCCUGACUG-----UGCACCA-----CCCACCCAUGCAGCCUU------ .(((((.....((((((....)))))--).((((((..((.((...((((.......)))))).))..)-----)))))..-----.........)))))...------ ( -25.00, z-score = -1.39, R) >droSec1.super_1 1264419 91 - 14215200 AGUUGCCACAAAGUCUAUUUUUAGAC--UCGUGCAUCCUUUGGUUUGCCGUGCAGCUUGGCCCUGACUG-----UGCACCA-----CCCACCCAUGCAACCUU------ .(((((.....((((((....)))))--).((((((..((.((...((((.......)))))).))..)-----)))))..-----.........)))))...------ ( -25.00, z-score = -1.87, R) >droYak2.chr2L 16281588 89 - 22324452 AGUUGCCACAAGGUCUAUUUUUAGAC--UCGUGCAUCCUUUAGUUUGCCGUGCAGCUUGGCCCUGACUGGUUGUUG------------CGCCCAAGUAACCUU------ .(((((.....((((((....)))))--).(((((.(..((((((.((((.......))))...))))))..).))------------)))....)))))...------ ( -25.70, z-score = -1.42, R) >droEre2.scaffold_4929 7642359 92 + 26641161 AGUUGCCACAAAGUCUAUUUUUAGAC--UCGGGCAUCCUUUAGUUUGCCGUGCAGCUCGGCCCUGACUGGCUGUUGC---------ACCACCCAUGCAACCUU------ .(((((.....((((((....)))))--)..((((..(....)..))))(((((((..((((......)))))))))---------)).......)))))...------ ( -32.60, z-score = -3.16, R) >droAna3.scaffold_13266 10117214 107 - 19884421 AACUGAUAUGAAUUCUAUUUAUAGUCCUUCCUGCAUUCAGUUUUUUGCC-CAUGACAUCGUAUUGAACGCCCUCAACCACC-CAACCCCACCGAGCCAGCGACAACCUU ......(((((((...)))))))(((((....((.((((((....((..-.....))....)))))).)).(((.......-..........)))..)).)))...... ( -9.63, z-score = 0.20, R) >consensus AGUUGCCACAAAGUCUAUUUUUAGAC__UCGUGCAUCCUUUGGUUUGCCGUGCAGCUUGGCCCUGACUGGCUGUUGCACC______CCCACCCAUGCAACCUU______ .(((((......(((((....)))))......(((....((((((.((((.......))))...))))))....)))..................)))))......... (-11.39 = -12.37 + 0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:15 2011