| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,576,892 – 3,577,055 |
| Length | 163 |
| Max. P | 0.894352 |

| Location | 3,576,892 – 3,577,055 |
|---|---|
| Length | 163 |
| Sequences | 4 |
| Columns | 165 |
| Reading direction | forward |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.43521 |
| G+C content | 0.39588 |
| Mean single sequence MFE | -42.14 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
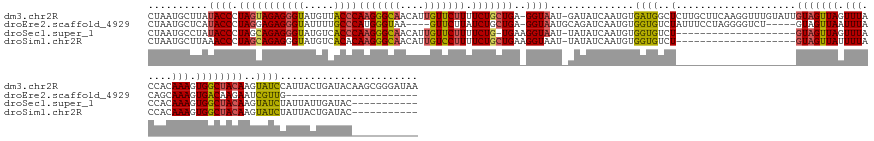
>dm3.chr2R 3576892 163 + 21146708 CUAAUGCUUAUACCCUAGUAGAGGGUAUGUUACCCAAGGGCAACAUUGUUCUUUUCUGCUGA-GGUAAU-GAUAUCAAUGUGAUGGCUCUUGCUUCAAGGUUUGUAUUGUAGUUAGUUUACCACAAAGUGGCUACAAGUAUCCAUUACUGAUACAAGCGGGAUAA ....(((((.((((.((((((((((((...)))))(((((((....))))))).))))))).-))))..-..(((((..(((((((.(((((...)))))......(((((((((.(((.....))).)))))))))....)))))))))))).)))))...... ( -49.60, z-score = -2.15, R) >droEre2.scaffold_4929 7610164 133 - 26641161 CUAAUGCUCAUACCCUAGGAGAGGGUAUUUUGCCCAUGGGUAA----GUUCUUAUCUGCUGA-GGUAAUGCAGAUCAAUGUGGUGUCUAUUUCCUAGGGGUCU-----GUAGUUAAUUUACAGCAAAGUGACAAGAAUCGUUG---------------------- ....((.((((.((((((((((((((.....)))).(((..(.----......((((((...-......))))))........)..)))))))))))))(.((-----((((.....)))))))...))))))..........---------------------- ( -40.06, z-score = -1.79, R) >droSec1.super_1 1232430 132 + 14215200 CUAAUGCCUAUACCCUAGCAGAGGGUAUGUCACCCAAGGGCAACAUUGUUCUUUUCUG-UGAAGGUAAU-UAUAUCAAUGUGGUGUCU--------------------GUAGUUAGUUUACCACAAAGUGGCUACAAGUAUCUAUUAUUGAUAC----------- ....((((((((((((.....))))))).((((..(((((((....)))))))....)-))))))))..-..((((((((.((((..(--------------------(((((((.(((.....))).))))))))..))))...)))))))).----------- ( -42.40, z-score = -3.29, R) >droSim1.chr2R 2290195 133 + 19596830 CUAAUGCUUAAACCCUAGCAGAGGGUAUGUCACACAAGGGCAACAUUGUCCUUUUCUGCUGAAGGUAAU-UAUAUCAAUGUGGUGUCU--------------------GUAGUUAUUUUACCACAAAGUGGCUACAAGUAUCUAUUACUGAUAC----------- ...........(((.(((((((((...((.....))((((((....)))))))))))))))..)))...-..(((((.((.((((..(--------------------(((((((((((.....))))))))))))..))))...)).))))).----------- ( -36.50, z-score = -1.69, R) >consensus CUAAUGCUUAUACCCUAGCAGAGGGUAUGUCACCCAAGGGCAACAUUGUUCUUUUCUGCUGA_GGUAAU_UAUAUCAAUGUGGUGUCU____________________GUAGUUAGUUUACCACAAAGUGGCUACAAGUAUCUAUUACUGAUAC___________ ...((((((.((((..((((((((((.....))))((((((......)))))).))))))...)))).........................................(((((((.(((.....))).)))))))))))))........................ (-20.27 = -21.53 + 1.25)


| Location | 3,576,892 – 3,577,055 |
|---|---|
| Length | 163 |
| Sequences | 4 |
| Columns | 165 |
| Reading direction | reverse |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.43521 |
| G+C content | 0.39588 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3576892 163 - 21146708 UUAUCCCGCUUGUAUCAGUAAUGGAUACUUGUAGCCACUUUGUGGUAAACUAACUACAAUACAAACCUUGAAGCAAGAGCCAUCACAUUGAUAUC-AUUACC-UCAGCAGAAAAGAACAAUGUUGCCCUUGGGUAACAUACCCUCUACUAGGGUAUAAGCAUUAG .......((((((((((((.((((...(((((.......(((((((......)))))))..((.....))..)))))..))))...)))))))..-..((((-..((.(((........((((((((....))))))))....))).))..)))))))))..... ( -41.00, z-score = -2.11, R) >droEre2.scaffold_4929 7610164 133 + 26641161 ----------------------CAACGAUUCUUGUCACUUUGCUGUAAAUUAACUAC-----AGACCCCUAGGAAAUAGACACCACAUUGAUCUGCAUUACC-UCAGCAGAUAAGAAC----UUACCCAUGGGCAAAAUACCCUCUCCUAGGGUAUGAGCAUUAG ----------------------....(((.((..(.......(((((.......)))-----))..((((((((................((((((......-...))))))......----........(((.......)))..)))))))).)..)).))).. ( -29.50, z-score = -1.63, R) >droSec1.super_1 1232430 132 - 14215200 -----------GUAUCAAUAAUAGAUACUUGUAGCCACUUUGUGGUAAACUAACUAC--------------------AGACACCACAUUGAUAUA-AUUACCUUCA-CAGAAAAGAACAAUGUUGCCCUUGGGUGACAUACCCUCUGCUAGGGUAUAGGCAUUAG -----------(((((.......))))).....(((.(..((((((...((......--------------------))..))))))..).....-..((((((..-((((........((((..((....))..))))....))))..))))))..)))..... ( -35.70, z-score = -2.55, R) >droSim1.chr2R 2290195 133 - 19596830 -----------GUAUCAGUAAUAGAUACUUGUAGCCACUUUGUGGUAAAAUAACUAC--------------------AGACACCACAUUGAUAUA-AUUACCUUCAGCAGAAAAGGACAAUGUUGCCCUUGUGUGACAUACCCUCUGCUAGGGUUUAAGCAUUAG -----------(((((.......))))).....((..(..((((((...........--------------------....))))))..).....-...(((((.((((((...((...((((..(......)..))))..)))))))))))))....))..... ( -31.16, z-score = -1.03, R) >consensus ___________GUAUCAGUAAUAGAUACUUGUAGCCACUUUGUGGUAAACUAACUAC____________________AGACACCACAUUGAUAUA_AUUACC_UCAGCAGAAAAGAACAAUGUUGCCCUUGGGUAACAUACCCUCUGCUAGGGUAUAAGCAUUAG .................................((..(..((((((...................................))))))..)...............................((((((....))))))(((((((.....)))))))..))..... (-15.65 = -16.52 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:11 2011