| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,568,330 – 3,568,470 |
| Length | 140 |
| Max. P | 0.609424 |

| Location | 3,568,330 – 3,568,470 |
|---|---|
| Length | 140 |
| Sequences | 12 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 70.37 |
| Shannon entropy | 0.66384 |
| G+C content | 0.42086 |
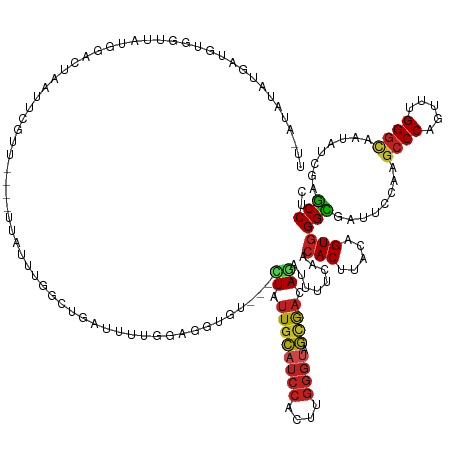
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
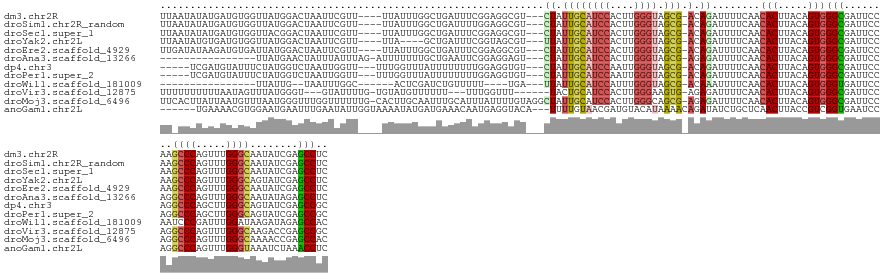
>dm3.chr2R 3568330 140 + 21146708 UUAAUAUAUGAUGUGGUUAUGGACUAAUUCGUU----UUAUUUGGCUGAUUUCGGAGGCGU---CUAUUGCAUCCACUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAAGCCCAGUUUGGGCAAUAUCGAGCCUC ..((((.((((..(((((...)))))..)))).----.)))).(((((((...((((.(((---(..((((((((....)))).)))-)...........(((.....))))))).))))..((((.....))))...))).)))).. ( -38.40, z-score = -0.92, R) >droSim1.chr2R_random 880671 140 + 2996586 UUAAUAUAUGAUGUGGUUAUGGACUAAUUCGUU----UUAUUUGGCUGAUUUUGGAGGCGU---CUAUUGCAUCCACUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAAGCCCAGUUUGGGCAAUAUCGAGCCUC ..((((.((((..(((((...)))))..)))).----.)))).(((((((.((((((.(((---(..((((((((....)))).)))-)...........(((.....))))))).))))))((((.....))))...))).)))).. ( -39.90, z-score = -1.40, R) >droSec1.super_1 1224045 140 + 14215200 UUAAUAUAUGAUGUGGUUACGGACUAAUUCGUU----UUAUUUGGCUGAUUUCGGAGGCGU---CUAUUGCAUCCACUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAAGCCCAGUUUGGGCAAUAUCGAGCCUC ..((((.((((..(((((...)))))..)))).----.)))).(((((((...((((.(((---(..((((((((....)))).)))-)...........(((.....))))))).))))..((((.....))))...))).)))).. ( -38.40, z-score = -0.97, R) >droYak2.chr2L 16239034 136 + 22324452 UUAAUAUGUGAUGUGGUUAUGGACUAAUUCGUU----UUA----GCUGAUUUCGGUAGCGU---UUAUUGCAUCCACUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAAGCCCAGUUUGGGCAGUAUCGAGCCUC ......(((((.((((((((.((..((((.((.----...----)).)))))).)))))((---((.((((((((....)))).)))-).))))......)))))))).(((((........)))))....((((........)))). ( -35.80, z-score = -0.41, R) >droEre2.scaffold_4929 7601757 140 - 26641161 UUGAUAUAAGAUGUGAUUAUGGACUAAUUCGUU----UUAUUUGGCUGAUUUCGGAGGCGU---CUAUUGCAUCCACUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAAGCCCAGUUUGGGCAAUAUCGAGCCUC .....((((((((.(((((.....)))))))))----))))..(((((((...((((.(((---(..((((((((....)))).)))-)...........(((.....))))))).))))..((((.....))))...))).)))).. ( -39.40, z-score = -1.25, R) >droAna3.scaffold_13266 10077274 127 + 19884421 ----------------UUAUGAACUAUUUAUUUAG-AUUUUUUUGCUGAAUUCGGAGGAGU---CUAUUGCAUCCACUUGGGUAGCG-AGAGAUUUUCAACACUUACAGUGGGCGAUUCCAGGCCCAGUUUGGGCAAUAUAGAGCCUC ----------------...((((..((((...(((-(((((((((.......)))))))))---)))((((((((....)))).)))-).)))).)))).(((.....)))(((........((((.....))))........))).. ( -33.39, z-score = -1.10, R) >dp4.chr3 13234504 136 - 19779522 -----UCGAUGUAUUUCUAUGGUCUAAUUGGUU---UUUGGUUUAUUUUUUUUGGAGGUGU---CUAUUGCAUCCAAUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAGGCCCAGCUUGGGCAGUAUCGAGCCGC -----((((((................((((..---.(((...........((((((..((---((.((((((((....)))).)))-).))))))))))(((.....)))..)))..))))((((.....))))..))))))..... ( -33.80, z-score = 0.23, R) >droPer1.super_2 2272057 136 - 9036312 -----UCGAUGUAUUUCUAUGGUCUAAUUGGUU---UUUGGUUUAUUUUUUUUGGAGGUGU---CUAUUGCAUCCAAUUGGGUAGCG-ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAGGCCCAGCUUGGGCAGUAUCGAGCCGC -----((((((................((((..---.(((...........((((((..((---((.((((((((....)))).)))-).))))))))))(((.....)))..)))..))))((((.....))))..))))))..... ( -33.80, z-score = 0.23, R) >droWil1.scaffold_181009 1200390 116 - 3585778 ----------------UUAUUG--UAAUUUGGC------ACUCGAUCUGUUUUU----UGA---UUAUUGCAUCCAUUUGGGUAGCG-ACAAAUUUUCAACACUUACAGUGGGUGAUUCCAAUCCCGAUUUGGAUAAGAUAGAGCCAC ----------------......--.....((((------..((((........)----)))---(((((..(((((.(((((.....-............(((.....)))((.....))...)))))..)))))..))))).)))). ( -25.00, z-score = -0.50, R) >droVir3.scaffold_12875 14618966 134 + 20611582 UUUUUUUUUUAAUAGUUUAUGGGU---GUAUUUUG-UGUAUGUUUUUU---UUUGGUUU------UACUGCAUCCACUUGGGAAGUG-AGAGAUUUUCAACACUUACAGUGGGCGAUUCCAGGCCCAGUUUGGGCAAGACCGAGCCGC .....................(((---((((....-.)))).......---.(((((((------(.((((..(((((((((...((-(((...)))))...)))).)))))..)....)))((((.....))))))))))))))).. ( -30.40, z-score = 0.10, R) >droMoj3.scaffold_6496 10098060 146 + 26866924 UUCACUUAUUAAUGUUUAAUGGGUUUGGUUUUUUG-CACUUGCAAUUUGCAUUUAUUUUGUAGGCUAUUGCAUCCACUUGGGCAGCG-AGAGAUUUUCAACACUUACAGUGGGCGAUUCCAGGCCCAGUUUGGGCAAAACCGAGCCAC ...................(((((((((....(((-(...(((((((((((.......)))))...)))))).(((((((((....(-(((...))))....)))).)))))))))..)))))))))((((((......))))))... ( -41.00, z-score = -1.39, R) >anoGam1.chr2L 3865506 139 + 48795086 ------UGAAAACGUGGAAUGAAUUUGAAUAUUGGUAAAAUAUGAUGAAACAAUGAGGUACA---UUUUGUAACGAUGUACAUAAAACAGAUAUCUGCUCAACUUACCGUCGGUGAAUCCAGGCCCAGUUUGGGUAAAUCUAAACCUC ------........((((.............((((.......(((((......(((((((((---((.......)))))))......(((....)))))))......)))))......))))((((.....))))...))))...... ( -26.72, z-score = 0.09, R) >consensus UU_AUAUAUGAUGUGGUUAUGGACUAAUUCGUU____UUAUUUGGCUGAUUUUGGAGGUGU___CUAUUGCAUCCACUUGGGUAGCG_ACAGAUUUUCAACACUUACAGUGGGCGAUUCCAAGCCCAGUUUGGGCAAUAUCGAGCCUC .....................................................................((((((....)))).))..............(((.....)))(((........((((.....))))........))).. (-17.25 = -17.50 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:09 2011