
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,556,527 – 3,556,650 |
| Length | 123 |
| Max. P | 0.641740 |

| Location | 3,556,527 – 3,556,650 |
|---|---|
| Length | 123 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 57.57 |
| Shannon entropy | 0.81563 |
| G+C content | 0.38599 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -9.03 |
| Energy contribution | -10.62 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
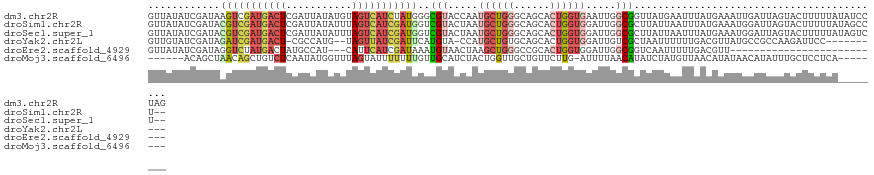
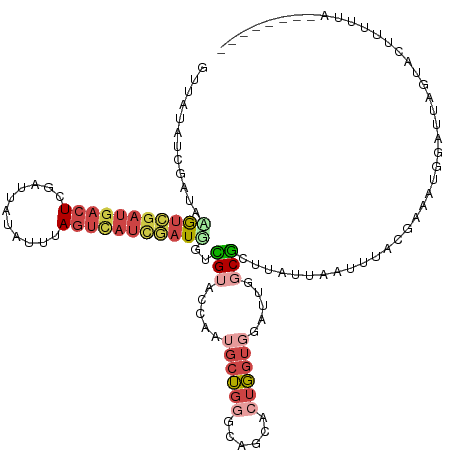
>dm3.chr2R 3556527 123 + 21146708 GUUAUAUCGAUAAGUCGAUGACUCGAUUAUAUGUAGUCAUCUAUGGGCGUACCAAUGCUGGGCAGCACUGGUGAAUUGGCGGUUAUGAAUUUAUGAAAUUGAUUAGUACUUUUUAUAUCCUAG ....((((((....))))))..((((((.((((...((((..((.(.(((((((.((((....)))).)))))...)).).)).))))...)))).))))))..................... ( -26.60, z-score = 0.23, R) >droSim1.chr2R 2283474 121 + 19596830 GUUAUAUCGAUACGUCGAUGACUCGAUUAUAUUUAGUCAUCGAUGGUCGUACUAAUGCUGGGCAGCACUGGUGGAUUGGCGCUUAUUAAUUUAUGAAAUGGAUUAGUACUUUUUAUAGCCU-- ((((((..(((.(((((((((((.(((...))).))))))))))))))(((((((((((....)))..(.(((((((((......))))))))).).....))))))))....))))))..-- ( -35.80, z-score = -2.88, R) >droSec1.super_1 1212673 121 + 14215200 GUUAUAUCGAUACGUCGAUGACUCGAUUAUAUUUAGUCAUCGAUGGUCGUACUAAUGCUGGGCAGCACUGGUGGAUUGGCGCUUAUUAAUUUAUGAAAUGGAUUAGUACUUUUUAUAGUCU-- .......((((.(((((((((((.(((...))).)))))))))))))))(((((.((((....)))).)))))(((((.....(((((((((((...))))))))))).......))))).-- ( -34.20, z-score = -2.52, R) >droYak2.chr2L 16227713 109 + 22324452 GUUGUAUCGAUAGAUCGAUGACU-CGCCAUG--UAGUUAUCGAUUCAUGUA-CCAUGCUGUGCAGCACUGGUGGAUUGUCGCUAAUUUUUUGACGUUAUGCCGCCAAGAUUCC---------- (((((((.(...(((((((((((-.......--.)))))))))))((((..-.))))).)))))))..((((((..(((((.........))))).....)))))).......---------- ( -30.70, z-score = -1.31, R) >droEre2.scaffold_4929 7590155 94 - 26641161 GUUAUAUCGAUAGGUCUAUGACUAUGCCAU---CAUUCAUCGAUAAAUGUAACUAAGCUGGGCCGCACUGGUGGAUUGGCGGUCAAUUUUUGACGUU-------------------------- (((((((..((.(((..((((........)---)))..))).))..)))))))...((..(.((((....)))).)..)).((((.....))))...-------------------------- ( -22.70, z-score = -0.38, R) >droMoj3.scaffold_6496 10086653 108 + 26866924 ------ACAGCUAACAGCUGUCUCAAUAUGGUUUAGUAUUUUUUUGUUGCAUCUACUGGUUGCUGUUCUUG-AUUUUAACAUAUCUAUGUUAACAUAUAACAUAUUUGCUCCUCA-------- ------(((((.....)))))........((...((((..(...(((((((((....)).)))........-...(((((((....))))))).....)))).)..))))))...-------- ( -15.40, z-score = 0.41, R) >consensus GUUAUAUCGAUAAGUCGAUGACUCGAUUAUAUUUAGUCAUCGAUGGUCGUACCAAUGCUGGGCAGCACUGGUGGAUUGGCGCUUAUUAAUUUACGAAAUGGAUUAGUACUUUUUA________ ............(((((((((((...........)))))))))))..(((.....((((((......)))))).....))).......................................... ( -9.03 = -10.62 + 1.59)
| Location | 3,556,527 – 3,556,650 |
|---|---|
| Length | 123 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 57.57 |
| Shannon entropy | 0.81563 |
| G+C content | 0.38599 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -5.07 |
| Energy contribution | -7.52 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
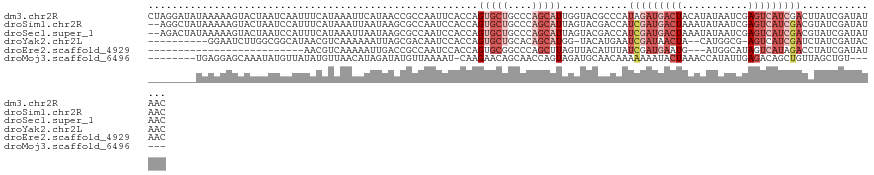
>dm3.chr2R 3556527 123 - 21146708 CUAGGAUAUAAAAAGUACUAAUCAAUUUCAUAAAUUCAUAACCGCCAAUUCACCAGUGCUGCCCAGCAUUGGUACGCCCAUAGAUGACUACAUAUAAUCGAGUCAUCGACUUAUCGAUAUAAC .....((((...((((...................................(((((((((....))))))))).........(((((((.(........)))))))).))))....))))... ( -24.30, z-score = -2.42, R) >droSim1.chr2R 2283474 121 - 19596830 --AGGCUAUAAAAAGUACUAAUCCAUUUCAUAAAUUAAUAAGCGCCAAUCCACCAGUGCUGCCCAGCAUUAGUACGACCAUCGAUGACUAAAUAUAAUCGAGUCAUCGACGUAUCGAUAUAAC --....((((....((((((((..................(((((..........)))))((...)))))))))).....(((((((((...........)))))))))........)))).. ( -23.60, z-score = -2.37, R) >droSec1.super_1 1212673 121 - 14215200 --AGACUAUAAAAAGUACUAAUCCAUUUCAUAAAUUAAUAAGCGCCAAUCCACCAGUGCUGCCCAGCAUUAGUACGACCAUCGAUGACUAAAUAUAAUCGAGUCAUCGACGUAUCGAUAUAAC --.((.(((.....((((((((..................(((((..........)))))((...)))))))))).....(((((((((...........))))))))).)))))........ ( -23.70, z-score = -3.03, R) >droYak2.chr2L 16227713 109 - 22324452 ----------GGAAUCUUGGCGGCAUAACGUCAAAAAAUUAGCGACAAUCCACCAGUGCUGCACAGCAUGG-UACAUGAAUCGAUAACUA--CAUGGCG-AGUCAUCGAUCUAUCGAUACAAC ----------.((.((((((((......)))))).......((........((((.((((....)))))))-).((((............--)))))))-).))(((((....)))))..... ( -24.50, z-score = -0.70, R) >droEre2.scaffold_4929 7590155 94 + 26641161 --------------------------AACGUCAAAAAUUGACCGCCAAUCCACCAGUGCGGCCCAGCUUAGUUACAUUUAUCGAUGAAUG---AUGGCAUAGUCAUAGACCUAUCGAUAUAAC --------------------------(((((((.....)))).(((.((......))..)))........))).....((((((((....---(((((...))))).....)))))))).... ( -16.00, z-score = -0.14, R) >droMoj3.scaffold_6496 10086653 108 - 26866924 --------UGAGGAGCAAAUAUGUUAUAUGUUAACAUAGAUAUGUUAAAAU-CAAGAACAGCAACCAGUAGAUGCAACAAAAAAAUACUAAACCAUAUUGAGACAGCUGUUAGCUGU------ --------.....(((.((((((((.((((....)))))))))))).....-....((((((..((((((.........................))))).)...)))))).)))..------ ( -17.11, z-score = -0.49, R) >consensus ________UAAAAAGUACUAAUCCAUUACAUAAAUAAAUAAGCGCCAAUCCACCAGUGCUGCCCAGCAUUGGUACAACCAUCGAUGACUAAAUAUAAUCGAGUCAUCGACCUAUCGAUAUAAC .......................................................(((((....)))))...........(((((((((...........))))))))).............. ( -5.07 = -7.52 + 2.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:06:07 2011