
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,287,927 – 2,287,986 |
| Length | 59 |
| Max. P | 0.988628 |

| Location | 2,287,927 – 2,287,986 |
|---|---|
| Length | 59 |
| Sequences | 7 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 72.70 |
| Shannon entropy | 0.54790 |
| G+C content | 0.49500 |
| Mean single sequence MFE | -15.29 |
| Consensus MFE | -6.34 |
| Energy contribution | -6.77 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
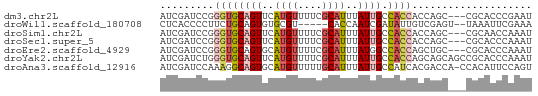
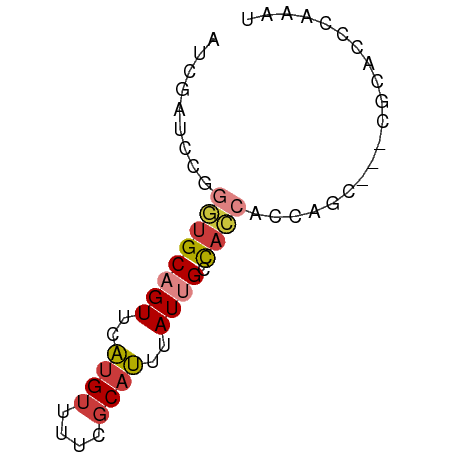
>dm3.chr2L 2287927 59 + 23011544 AUCGAUCCGGGUGCAGUUCAUGUUUUCGCAUUUAUUGCCACCACCAGC---CGCACCCGAAU .......(((((((.(((..((.....(((.....)))...))..)))---.)))))))... ( -18.10, z-score = -3.12, R) >droWil1.scaffold_180708 2252056 55 + 12563649 CUCACCCCUUCUGCAGUGUGCGU-----CACCAAUCGAUAUUGUCGAGU--UAAAUUCGAAA ........(((.(((((((.((.-----.......))))))))).))).--........... ( -8.00, z-score = -0.55, R) >droSim1.chr2L 2248341 59 + 22036055 AUCGAUCCGGGUGCAGUUCAUGUUUUCGCAUUUAUUGCCACCACCAGC---CGCAACCAAAU .........((((((((..((((....))))..)))).))))......---........... ( -11.30, z-score = -0.61, R) >droSec1.super_5 462539 59 + 5866729 AUCGAUCCGGGUGCAGUUCAUGUUUUCGCAUUUAUUGCCACCACCAGC---CGCACCCAAAU ........((((((.(((..((.....(((.....)))...))..)))---.)))))).... ( -16.30, z-score = -2.85, R) >droEre2.scaffold_4929 2324861 59 + 26641161 AUCGAUCCGGGUGCAGUGCAUGUUUUCGCAUUUAUGGCCACCAGCUGC---CGCACCCAAAU ........(((((((((((........)))))...(((........))---))))))).... ( -20.60, z-score = -1.99, R) >droYak2.chr2L 2269306 62 + 22324452 AUCGAUCUGGGUGCAGUUCAUGUUUUCGCAUUUAUUGCCACCAGCAGCAGCCGCACCCAAAU .......(((((((.(((..((((...(((.....)))....))))..))).)))))))... ( -19.80, z-score = -2.97, R) >droAna3.scaffold_12916 10001554 61 - 16180835 AUCGAUCCAAAGGCAGUGCAUGUUUUUGCAUUUAUUGCCAUCACGACCA-CCACAUUCCAGU ...........(((((((.((((....)))).)))))))..........-............ ( -12.90, z-score = -2.02, R) >consensus AUCGAUCCGGGUGCAGUUCAUGUUUUCGCAUUUAUUGCCACCACCAGC___CGCACCCAAAU .........((((((((..(((......)))..)))).)))).................... ( -6.34 = -6.77 + 0.43)
| Location | 2,287,927 – 2,287,986 |
|---|---|
| Length | 59 |
| Sequences | 7 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 72.70 |
| Shannon entropy | 0.54790 |
| G+C content | 0.49500 |
| Mean single sequence MFE | -19.19 |
| Consensus MFE | -10.27 |
| Energy contribution | -10.34 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
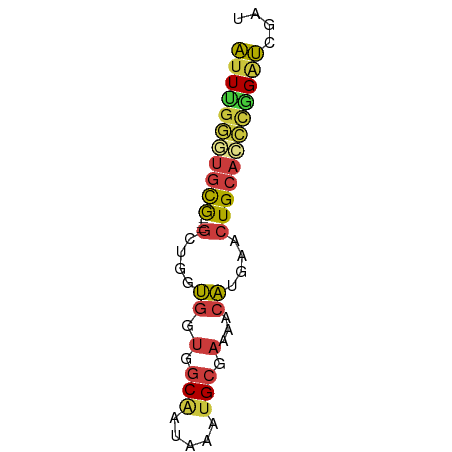
>dm3.chr2L 2287927 59 - 23011544 AUUCGGGUGCG---GCUGGUGGUGGCAAUAAAUGCGAAAACAUGAACUGCACCCGGAUCGAU (((((((((((---(.(.(((.(.(((.....))).)...))).).)))))))))))).... ( -26.50, z-score = -4.49, R) >droWil1.scaffold_180708 2252056 55 - 12563649 UUUCGAAUUUA--ACUCGACAAUAUCGAUUGGUG-----ACGCACACUGCAGAAGGGGUGAG ..((((.....--..))))...........(...-----.)...((((.(....).)))).. ( -8.40, z-score = 0.26, R) >droSim1.chr2L 2248341 59 - 22036055 AUUUGGUUGCG---GCUGGUGGUGGCAAUAAAUGCGAAAACAUGAACUGCACCCGGAUCGAU ((((((.((((---(.(.(((.(.(((.....))).)...))).).))))).)))))).... ( -17.90, z-score = -1.56, R) >droSec1.super_5 462539 59 - 5866729 AUUUGGGUGCG---GCUGGUGGUGGCAAUAAAUGCGAAAACAUGAACUGCACCCGGAUCGAU (((((((((((---(.(.(((.(.(((.....))).)...))).).)))))))))))).... ( -24.50, z-score = -4.04, R) >droEre2.scaffold_4929 2324861 59 - 26641161 AUUUGGGUGCG---GCAGCUGGUGGCCAUAAAUGCGAAAACAUGCACUGCACCCGGAUCGAU (((((((((((---(..((((.(.((.......)).)...)).)).)))))))))))).... ( -21.50, z-score = -1.57, R) >droYak2.chr2L 2269306 62 - 22324452 AUUUGGGUGCGGCUGCUGCUGGUGGCAAUAAAUGCGAAAACAUGAACUGCACCCAGAUCGAU ((((((((((((.(((((....)))))....(((......)))...)))))))))))).... ( -21.80, z-score = -2.52, R) >droAna3.scaffold_12916 10001554 61 + 16180835 ACUGGAAUGUGG-UGGUCGUGAUGGCAAUAAAUGCAAAAACAUGCACUGCCUUUGGAUCGAU .(..((....((-..((((((...(((.....))).....)))).))..))))..)...... ( -13.70, z-score = -0.53, R) >consensus AUUUGGGUGCG___GCUGGUGGUGGCAAUAAAUGCGAAAACAUGAACUGCACCCGGAUCGAU (((((((((((...........(.(((.....))).)..........))))))))))).... (-10.27 = -10.34 + 0.07)

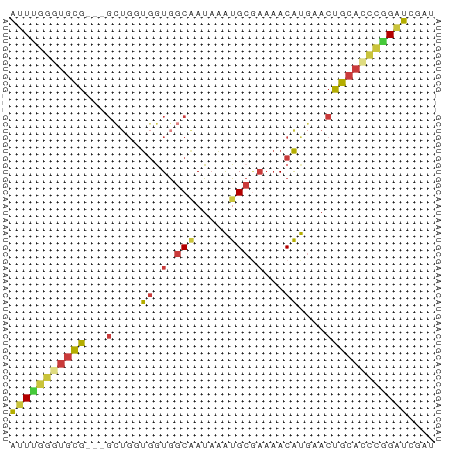
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:46 2011