| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,481,236 – 3,481,332 |
| Length | 96 |
| Max. P | 0.968533 |

| Location | 3,481,236 – 3,481,332 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 54.79 |
| Shannon entropy | 0.91301 |
| G+C content | 0.46483 |
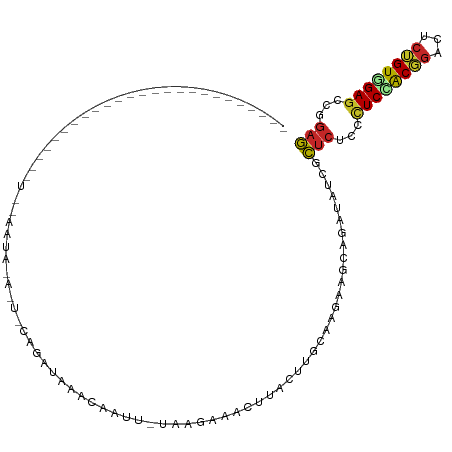
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -8.81 |
| Energy contribution | -8.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
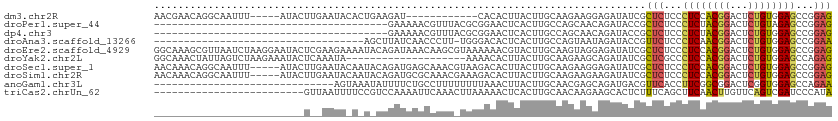
>dm3.chr2R 3481236 96 - 21146708 AACGAACAGGCAAUUU-----AUACUUGAAUACACUGAAGAU------------CACACUUACUUGCAAGAAGGAGAUAUCGCUCUCCCUCCACGGACUCUGUGGAGCCGGAG ..(((.(((...((((-----(....)))))...)))...((------------(.(.(((.((....)))))).))).)))..((((((((((((...))))))))..)))) ( -22.30, z-score = -1.23, R) >droPer1.super_44 169810 74 + 614636 ---------------------------------------GAAAAACGUUUACGCGGAACUCACUUGCCAGCAACAGAUACCGCUCUCCCUCUACGGACUCUGUAGAGCCGGAG ---------------------------------------.............((((...((..(((....)))..))..)))).((((((((((((...))))))))..)))) ( -20.50, z-score = -2.00, R) >dp4.chr3 7518250 74 + 19779522 ---------------------------------------GAAAAACGUUUACGCGGAACUCACUUGCCAGCAACAGAUACCGCUCUCCCUCUACGGACUCUGUGGAGCCGGAG ---------------------------------------.............((((...((..(((....)))..))..)))).((((((((((((...))))))))..)))) ( -20.20, z-score = -1.43, R) >droAna3.scaffold_13266 10001818 77 - 19884421 -----------------------------------AGCUUAUCAACCCUU-UGGGACACUCACUUGCCAGUAAUAGAUACCGUUCUCCCUCAACGGACUCUGUGGAGCCGGAA -----------------------------------.((((.....(((..-.))).................(((((..(((((.......)))))..))))).))))..... ( -14.30, z-score = 1.07, R) >droEre2.scaffold_4929 7524559 113 + 26641161 GGCAAAGCGUUAAUCUAAGGAAUACUCGAAGAAAAUACAGAUAAACAAGCGUAAAAAACGUACUUGCAAGUAGGAGAUAUCGCUCUCCCUCCACGGACUCUGUGGAGCCGGAG (((...(((((.((((......................)))).)))..((((.....))))....)).....(((((......))))).(((((((...)))))))))).... ( -26.45, z-score = -0.86, R) >droYak2.chr2L 16159140 93 - 22324452 GGCAAACUAUUAGUCUAAGAAAUACUCAAAUA--------------------AAAACACUUACUUGCAAGAAGCAGAUAUCGCUCGCCCUCCACGGACUCUGUGGAGCCAGAG (((...............((.....)).....--------------------...........((((.....)))).........)))((((((((...))))))))...... ( -17.10, z-score = -0.37, R) >droSec1.super_1 1144278 108 - 14215200 AACAAACAGGCAAUUU-----AUACUUGAAUACAAUACAGAUGAGCAAACGUAAGACACUUACUUGCAAGAAGGAGAUAUCGCUCUCCCUCCACGGACUCUGUGGAGCCGGAG ........(((.....-----.......................((((..(((((...))))))))).....(((((......))))).(((((((...)))))))))).... ( -25.60, z-score = -1.84, R) >droSim1.chr2R 2211698 108 - 19596830 AACAAACAGGCAAUUU-----AUACUUGAAUACAAUACAGAUGCGCAAACGAAAGACACUUACUUGCAAGAAGAAGAUAUCGCUCUCCCUCCACGGACUCUGUGGAGCCGGAG ........(((.((((-----(....)))))........((((......(....)...(((.(((.....)))))).)))))))((((((((((((...))))))))..)))) ( -22.70, z-score = -1.46, R) >anoGam1.chr3L 33785076 83 + 41284009 ------------------------------AGUAAAUAUUUUCUGCCUUUUUUUUAAACUUACUUGCAACGAGCAGAUGACGUUCACCUUCGGCGGACUCGGUGGAGCCAGAA ------------------------------..........(((..((................((((.....))))..((.((((.((...)).))))))))..)))...... ( -15.60, z-score = 0.57, R) >triCas2.chrUn_62 126849 88 - 341982 -------------------------GUUAAUUUUCCGUCCAAAAUUCAAACUUAAAAACUCACUUGCAACAAGAAGCACUCUUUCAGCUUCAACUUGUUCAGUCGAUCCCAUA -------------------------..................................(((((.((((...(((((.........)))))...))))..))).))....... ( -8.70, z-score = -1.07, R) >consensus _________________________U__AAUA_A_U_CAGAUAAACAAUU_UAAGAAACUUACUUGCAAGAAGCAGAUAUCGCUCUCCCUCCACGGACUCUGUGGAGCCGGAG ..................................................................................(((...((((((((...))))))))...))) ( -8.81 = -8.70 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:58 2011