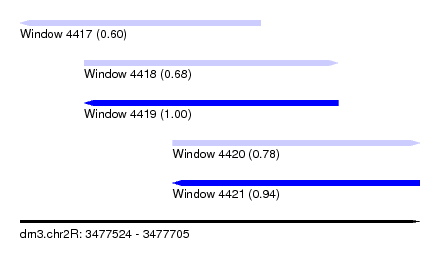
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,477,524 – 3,477,705 |
| Length | 181 |
| Max. P | 0.998833 |

| Location | 3,477,524 – 3,477,633 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Shannon entropy | 0.64716 |
| G+C content | 0.43752 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -10.66 |
| Energy contribution | -12.99 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
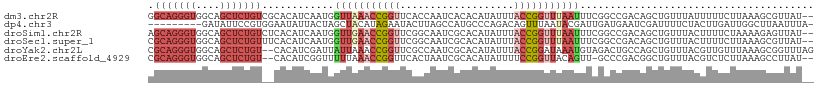
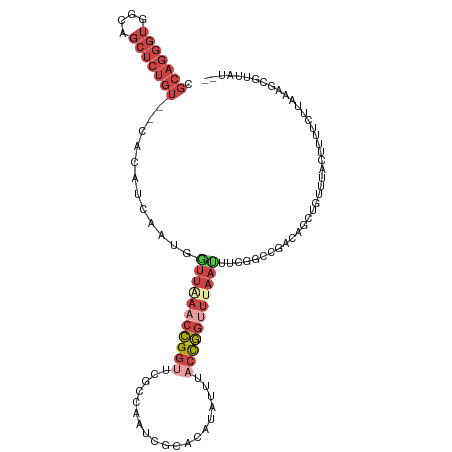
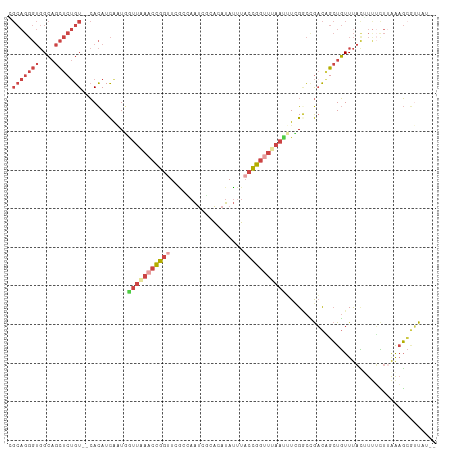
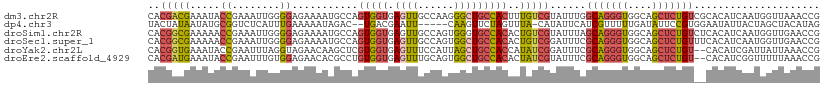
>dm3.chr2R 3477524 109 - 21146708 GGCAGGGUGGCAGCUCUGUCGCACAUCAAUGGUUAAACCGGUUCACCAAUCACACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUUUAUUUUUCUUAAAGCGUUAU-- ((((((((....))))))))((........((((((((((((...................)))))))))))).((((.....))))...............)).....-- ( -29.61, z-score = -2.53, R) >dp4.chr3 7514465 101 + 19779522 ---------GAUAUUCCGUGGAAUAUUACUAGCUACAUAGAAUACUUAGCCAUGCCCAGACAGUUUAAUACGAUUGAUGAAUCGAUUUUCUACUUGAUUGGCUUAAUUUA- ---------(((((((....))))))).(((......))).......(((((....(((..((...(((.(((((....))))))))..))..)))..))))).......- ( -17.10, z-score = -0.94, R) >droSim1.chr2R 2207896 109 - 19596830 AGCAGGGUGGCAGCUCUGUCUCACAUCAAUGGUUGAACCGGUUCGGCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUUUACUUUUCUAAAAGAGUUAU-- .(((((((....)))))))(((........((((((((((((...((....))........)))))))))))).((((.....))))...............)))....-- ( -27.90, z-score = -1.04, R) >droSec1.super_1 1140570 109 - 14215200 CGCAGGGUGGCAGCUCUGUUUCACAUCAAUGGUUGAACCGGUUCGGCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUUUACUUUUCUUAAAGCGUUAU-- .(((((((....)))))))........(((((((((((((((...((....))........)))))))))))..((((.....))))................))))..-- ( -26.20, z-score = -0.43, R) >droYak2.chr2L 16155417 109 - 22324452 CGCAGGGUGGCAGCUCUGU--CACAUCGAUUAUUAAACCGGUUCGCCAAUCGCACAUAUUUACCGGAUAAAUGUAGACUGCCAGCUGUUUACGUUGUUUAAAGCGGUUUAG (((...((((((....)))--)))........((((((((((...................)))))...(((((((((........))))))))).))))).)))...... ( -26.91, z-score = -0.25, R) >droEre2.scaffold_4929 7520839 106 + 26641161 CGCAGGGUGGCAGCUCUGU--CACAUCGGUUUUUAAACCGGUUCACUAAUCGCACAUAUUUUCCGGUUACAGUU-GCCCGACGGCUGUUUACGUCUCUUAAAGCCUUAU-- ...((((((((((((....--...(((((((....)))))))....((((((...........)))))).))))-))).((((........)))).......)))))..-- ( -28.30, z-score = -1.53, R) >consensus CGCAGGGUGGCAGCUCUGU__CACAUCAAUGGUUAAACCGGUUCGCCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUUUACUUUUCUUAAAGCGUUAU__ .(((((((....)))))))............(((((((((((...................)))))))))))....................................... (-10.66 = -12.99 + 2.34)
| Location | 3,477,553 – 3,477,668 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.03 |
| Shannon entropy | 0.58070 |
| G+C content | 0.47807 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -12.58 |
| Energy contribution | -13.67 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.677981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3477553 115 + 21146708 CGGCCGAAAUUAAACCGGUAAAUAUGUGUGAUUGGUGAACCGGUUUAACCAUUGAUGUGCGACAGAGCUGCCACCCUGCCAAAUACGACAAAGUGGCAGCCCUUGGCAACUCACC ..(((((..((((((((((..((...........))..))))))))))................(.((((((((..((.(......).))..)))))))).))))))........ ( -34.00, z-score = -1.52, R) >dp4.chr3 7514499 105 - 19779522 CAAUCGUA-UUAAACUGUC---UGGGCAUGGCUAAGUAUUCUAUGUAGCUAGUAAUAUUCCACGGAAUAUCAAAAACGAUGAAUAUG-UAAACUAGAACUUG-----AAUUCGUC ....((.(-((......((---(((((.((((((.(((...))).)))))))).((((((....)))))).................-....))))).....-----))).)).. ( -16.60, z-score = 0.12, R) >droSim1.chr2R 2207925 115 + 19596830 CGGCCGAAAUUAAACCGGUAAAUAUGUGCGAUUGCCGAACCGGUUCAACCAUUGAUGUGAGACAGAGCUGCCACCCUGCUAAAUACGACAGUGUGGCACCCACUGGCAACUCACC ..((((.........))))......(((.(.((((((....(((((..((((....))).)...)))))(((((.((((.......).))).)))))......))))))).))). ( -29.80, z-score = -0.64, R) >droSec1.super_1 1140599 115 + 14215200 CGGCCGAAAUUAAACCGGUAAAUAUGUGCGAUUGCCGAACCGGUUCAACCAUUGAUGUGAAACAGAGCUGCCACCCUGCGAAAUCCGACAGUGUGGCAGCCACUGGCAACUCACC ..((((.........))))......(((.(.((((((....((.....))..............(.((((((((.(((((.....)).))).)))))))))..))))))).))). ( -37.50, z-score = -2.54, R) >droYak2.chr2L 16155448 113 + 22324452 CAGUCUACAUUUAUCCGGUAAAUAUGUGCGAUUGGCGAACCGGUUUAAUAAUCGAUGUG--ACAGAGCUGCCACCCUGCGAAAUCCGAUAUGGUGGCAGCUAAUGGAAACUCACC ((((((((((.............))))).)))))......(((((....)))))..(((--(...((((((((((.((((.....)).)).))))))))))...(....))))). ( -34.82, z-score = -2.37, R) >droEre2.scaffold_4929 7520868 112 - 26641161 CGGGCAAC-UGUAACCGGAAAAUAUGUGCGAUUAGUGAACCGGUUUAAAAACCGAUGUG--ACAGAGCUGCCACCCUGCGAAAUACGAUAGUGUGGCAGCCACUGCAAACUCACC (((....)-)).......................((((..(((((....)))))...((--.(((.((((((((.(((((.....)).))).))))))))..)))))...)))). ( -33.90, z-score = -2.36, R) >consensus CGGCCGAAAUUAAACCGGUAAAUAUGUGCGAUUGGCGAACCGGUUUAACCAUUGAUGUG__ACAGAGCUGCCACCCUGCGAAAUACGACAGUGUGGCAGCCACUGGCAACUCACC ............(((((((...((........))....))))))).....................((((((((..................))))))))............... (-12.58 = -13.67 + 1.09)


| Location | 3,477,553 – 3,477,668 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.03 |
| Shannon entropy | 0.58070 |
| G+C content | 0.47807 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.998833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
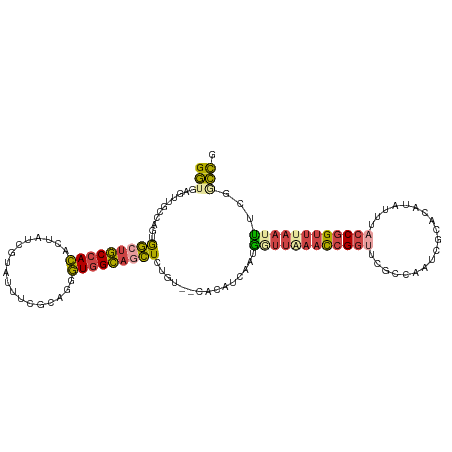
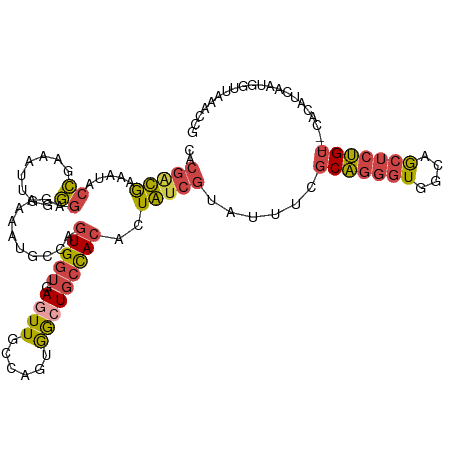
>dm3.chr2R 3477553 115 - 21146708 GGUGAGUUGCCAAGGGCUGCCACUUUGUCGUAUUUGGCAGGGUGGCAGCUCUGUCGCACAUCAAUGGUUAAACCGGUUCACCAAUCACACAUAUUUACCGGUUUAAUUUCGGCCG (((((..(((..((((((((((((((((((....))))))))))))))))))...)))..))...((((((((((((...................))))))))))))...))). ( -52.21, z-score = -6.29, R) >dp4.chr3 7514499 105 + 19779522 GACGAAUU-----CAAGUUCUAGUUUA-CAUAUUCAUCGUUUUUGAUAUUCCGUGGAAUAUUACUAGCUACAUAGAAUACUUAGCCAUGCCCA---GACAGUUUAA-UACGAUUG ........-----...(((((((....-).........(((..((((((((....))))))))..)))....))))))...............---..(((((...-...))))) ( -12.80, z-score = 0.67, R) >droSim1.chr2R 2207925 115 - 19596830 GGUGAGUUGCCAGUGGGUGCCACACUGUCGUAUUUAGCAGGGUGGCAGCUCUGUCUCACAUCAAUGGUUGAACCGGUUCGGCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCG .(((((....(((.((.((((((.((((........)))).)))))).))))).)))))......((((((((((((...((....))........))))))......)))))). ( -42.00, z-score = -2.56, R) >droSec1.super_1 1140599 115 - 14215200 GGUGAGUUGCCAGUGGCUGCCACACUGUCGGAUUUCGCAGGGUGGCAGCUCUGUUUCACAUCAAUGGUUGAACCGGUUCGGCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCG .(((((....(((.(((((((((.(((.((.....))))).)))))))))))).)))))......((((((((((((...((....))........))))))......)))))). ( -44.80, z-score = -3.14, R) >droYak2.chr2L 16155448 113 - 22324452 GGUGAGUUUCCAUUAGCUGCCACCAUAUCGGAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGAUUAUUAAACCGGUUCGCCAAUCGCACAUAUUUACCGGAUAAAUGUAGACUG ....(((((.((((((((((((((....((.....))...))))))))))....--................(((((...................)))))...)))).))))). ( -32.31, z-score = -1.81, R) >droEre2.scaffold_4929 7520868 112 + 26641161 GGUGAGUUUGCAGUGGCUGCCACACUAUCGUAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGGUUUUUAAACCGGUUCACUAAUCGCACAUAUUUUCCGGUUACA-GUUGCCCG (((((((..((((.(((((((((.((..((.....))..)))))))))))))))--.))(((((((....)))))))))))).................(((....-...))).. ( -35.90, z-score = -2.51, R) >consensus GGUGAGUUGCCAGUGGCUGCCACACUAUCGUAUUUCGCAGGGUGGCAGCUCUGU__CACAUCAAUGGUUAAACCGGUUCGCCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCG (((((((.......(((((((((..................))))))))).......)).))...((((((((((((...................))))))))))))...))). (-20.14 = -20.24 + 0.09)


| Location | 3,477,593 – 3,477,705 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.57142 |
| G+C content | 0.47680 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.77 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3477593 112 + 21146708 CGGUUUAACCAUUGAUGUGCGACAGAGCUGCCACCCUGCCAAAUACGACAAAGUGGCAGCCCUUGGCAACUCACCACUGGCAUUUUCUCCCCAAUUUCGGUAUUUCGUCGUG .((.....))........(((((.(((.(((((..((((((............))))))....))))).)))...(((((.(((........))).))))).....))))). ( -30.50, z-score = -1.19, R) >dp4.chr3 7514535 104 - 19779522 CUAUGUAGCUAGUAAUAUUCCACGGAAUAUCAAAAACGAUGAAUAUG-UAAACUAGAACUUG-----AAUUCGUCA--GUCUAUUUUUCAAAUGAGACCGCAUAUUAUAGUA (((((((((.....((((((....)))))).......(((((((..(-(........))...-----.))))))).--(((((((.....))).)))).)).)).))))).. ( -14.80, z-score = 0.27, R) >droSim1.chr2R 2207965 112 + 19596830 CGGUUCAACCAUUGAUGUGAGACAGAGCUGCCACCCUGCUAAAUACGACAGUGUGGCACCCACUGGCAACUCACCACUGGCAUUUUCUCCCCAAUUUCGGUUUUUCGCCGUG ((((..((((......(((((.(((.(.((((((.((((.......).))).)))))).)..)))....)))))...(((..........))).....))))....)))).. ( -28.50, z-score = -0.44, R) >droSec1.super_1 1140639 112 + 14215200 CGGUUCAACCAUUGAUGUGAAACAGAGCUGCCACCCUGCGAAAUCCGACAGUGUGGCAGCCACUGGCAACUCACCACUGGCAUUUUCUCCCCAAUUUCGGUUUUUCGCCGUG ((((..((((((((..(.((((......((((((.(((((.....)).))).))))))((((.(((.......))).))))..)))).)..))))...))))....)))).. ( -34.90, z-score = -1.89, R) >droYak2.chr2L 16155488 110 + 22324452 CGGUUUAAUAAUCGAUGUG--ACAGAGCUGCCACCCUGCGAAAUCCGAUAUGGUGGCAGCUAAUGGAAACUCACCACGAGCUUGUUCUACCUAAAUUCGGUAUUUCACCGUG .(((..((((((((..(((--(...((((((((((.((((.....)).)).))))))))))...(....)))))..)))..)))))..)))......((((.....)))).. ( -36.60, z-score = -3.74, R) >droEre2.scaffold_4929 7520907 110 - 26641161 CGGUUUAAAAACCGAUGUG--ACAGAGCUGCCACCCUGCGAAAUACGAUAGUGUGGCAGCCACUGCAAACUCACCACAGGCGUGUUCUCCACAAAUUCGGUAUUUCAUCGUG ((((..(((.(((((((((--.(((.((((((((.(((((.....)).))).))))))))..)))..(((.(.((...)).).)))...))))...))))).))).)))).. ( -33.70, z-score = -2.16, R) >consensus CGGUUUAACCAUUGAUGUG__ACAGAGCUGCCACCCUGCGAAAUACGACAGUGUGGCAGCCACUGGCAACUCACCACUGGCAUUUUCUCCCCAAAUUCGGUAUUUCACCGUG ((((......(((((..((....(((((((((((..................))))))))....(....)...............)))...))...))))).....)))).. (-13.18 = -12.77 + -0.41)

| Location | 3,477,593 – 3,477,705 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.57142 |
| G+C content | 0.47680 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3477593 112 - 21146708 CACGACGAAAUACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAAGGGCUGCCACUUUGUCGUAUUUGGCAGGGUGGCAGCUCUGUCGCACAUCAAUGGUUAAACCG ............(((....)))((.......((((.(((((...(((..((((((((((((((((((....))))))))))))))))))...)))))))).))))....)). ( -45.80, z-score = -3.74, R) >dp4.chr3 7514535 104 + 19779522 UACUAUAAUAUGCGGUCUCAUUUGAAAAAUAGAC--UGACGAAUU-----CAAGUUCUAGUUUA-CAUAUUCAUCGUUUUUGAUAUUCCGUGGAAUAUUACUAGCUACAUAG ..((((((((((((((((...((....)).))))--))..((((.-----...)))).......-))))))....(((..((((((((....))))))))..)))...)))) ( -20.40, z-score = -1.64, R) >droSim1.chr2R 2207965 112 - 19596830 CACGGCGAAAAACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAGUGGGUGCCACACUGUCGUAUUUAGCAGGGUGGCAGCUCUGUCUCACAUCAAUGGUUGAACCG ..(((.....((((...(((((.((((.....(((.((((.....)))).))).((((((.((((........)))).))))))......))))...)))))))))...))) ( -38.00, z-score = -1.16, R) >droSec1.super_1 1140639 112 - 14215200 CACGGCGAAAAACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAGUGGCUGCCACACUGUCGGAUUUCGCAGGGUGGCAGCUCUGUUUCACAUCAAUGGUUGAACCG ..(((.....(((((..(((((.(......).))))).(((((....(((.(((((((((.(((.((.....))))).)))))))))))).))))).....)))))...))) ( -40.10, z-score = -1.43, R) >droYak2.chr2L 16155488 110 - 22324452 CACGGUGAAAUACCGAAUUUAGGUAGAACAAGCUCGUGGUGAGUUUCCAUUAGCUGCCACCAUAUCGGAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGAUUAUUAAACCG ..(((((...((((.......))))((..((((((.....)))))).....((((((((((....((.....))...))))))))))...)--).)))))............ ( -34.00, z-score = -2.42, R) >droEre2.scaffold_4929 7520907 110 + 26641161 CACGAUGAAAUACCGAAUUUGUGGAGAACACGCCUGUGGUGAGUUUGCAGUGGCUGCCACACUAUCGUAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGGUUUUUAAACCG .....(((((.(((((...((((..((((.((((...)))).))))((((.(((((((((.((..((.....))..)))))))))))))))--))))))))))))))..... ( -39.40, z-score = -2.55, R) >consensus CACGACGAAAUACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAGUGGCUGCCACACUAUCGUAUUUCGCAGGGUGGCAGCUCUGU__CACAUCAAUGGUUAAACCG ..(((((.....((........))...........(((((.((((......)))))))))..)))))......(((((((....)))))))..................... (-14.21 = -14.38 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:57 2011