| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,476,739 – 3,476,830 |
| Length | 91 |
| Max. P | 0.737161 |

| Location | 3,476,739 – 3,476,830 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 67.88 |
| Shannon entropy | 0.70010 |
| G+C content | 0.52599 |
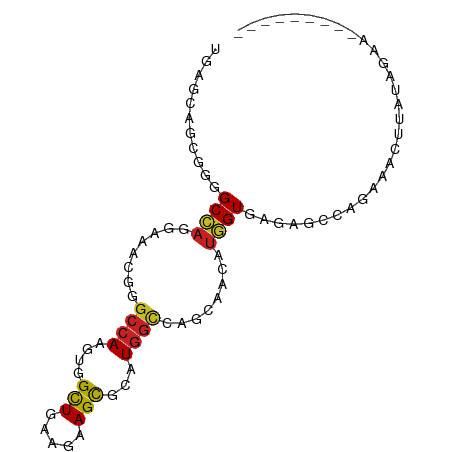
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -9.84 |
| Energy contribution | -9.79 |
| Covariance contribution | -0.05 |
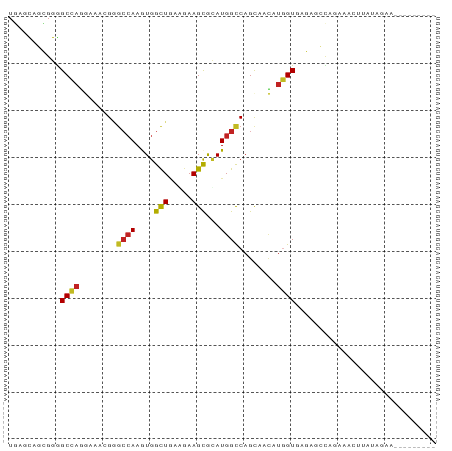
| Combinations/Pair | 1.36 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
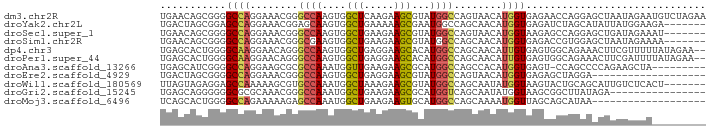
>dm3.chr2R 3476739 91 - 21146708 UGAACAGCGGGGCCAGGAAACGGGCCAAGUGGCUCAAGAAGCGUAUGGCCAGUAACAUGGUGAGAACCAGGAGCUAAUAGAAUGUCUAGAA .....(((..((((((....)(((((....)))))..........))))).......((((....))))...)))..(((.....)))... ( -27.80, z-score = -1.95, R) >droYak2.chr2L 16154576 84 - 22324452 UGACUAGCGGAGCCAGGAAACGGAGCAAGUGGCUGAAAAAGCGAAUGGCCAGCAACAUGGUGAGAUCUAGCAUAUUAUGGAAGA------- ......((.(.(((((....)..........(((.....)))...))))).))..(((((((.(......).))))))).....------- ( -17.60, z-score = 0.00, R) >droSec1.super_1 1139777 84 - 14215200 UGAACAGCGGGGCCAGGAAACGGGCCAAGUGGCUGAAGAAGCGUAUGGCCAGUAACAUGGUAAGAGCCAGGAGCUGAUAGAAAU------- ....((((..((((((....).((((....))))...........))))).......((((....))))...))))........------- ( -27.00, z-score = -2.69, R) >droSim1.chr2R 2207100 84 - 19596830 UGAACAGCGGGGCCAGGAAACGGGCGAAGUGGCUGAAGAAGCGUAUGGCCAGCAACAUGGUGAGACCGUGGAGCUAAUAGAAAA------- ..........((((((....)..........(((.....)))...)))))(((..(((((.....)))))..))).........------- ( -25.30, z-score = -2.06, R) >dp4.chr3 7513803 89 + 19779522 UGAGCACUGGGGCAAGGAACAGGGCCAAGUGGCUGAGGAAGCACAUGGCCAGCAACAUUGUGAGUGGCAGAAACUUCGUUUUUAUAGAA-- ....((((...((((.......(((((.(((.((.....))))).))))).......)))).))))..((((((...))))))......-- ( -21.44, z-score = 0.53, R) >droPer1.super_44 165340 89 + 614636 UGAGCACUGGGGCAAGGAACAGGGCCAAGUGGCUGAGGAAGCACAUGGCCAGCAACAUUGUGAGUGGCAGAAACUUCGAUUUUAUAGAA-- ...((.(((..........)))(((((.(((.((.....))))).))))).))...((((.((((.......)))))))).........-- ( -21.50, z-score = 0.16, R) >droAna3.scaffold_13266 9997511 81 - 19884421 UGAGCAUCGGGGCCAGGAAGCGCGCCAAAUGGUUGAAGAAGCGCAUGGCCAGCCACAUGGUGAGU-CCAGCCCCAGAAGCUA--------- ..(((.(((((((..(((....(((((..((((((.....((.....))))))))..)))))..)-)).))))).)).))).--------- ( -34.20, z-score = -2.58, R) >droEre2.scaffold_4929 7520066 72 + 26641161 UGACUAGCGGGGCCAGGAAACGGGCCAAGUGGCUGAGGAAGCGUAUGGCCAGUAACAUGGUGAGAGCUAGGA------------------- ...(((((..((((.(....).)))).....(((.....)))...(.((((......)))).)..)))))..------------------- ( -26.30, z-score = -2.77, R) >droWil1.scaffold_180569 1184112 84 + 1405142 UUAGUAGAGGAGCCAAAAAGCGUGCCAAAUGGCUAAAGAAGCGUAUGGCCAGCAAUAUGGUAAGUACUGCAGCAUUGUCUCACU------- .........(((.(((...((..(((....))).......((((((.((((......))))..)))).)).)).))).)))...------- ( -19.40, z-score = 0.32, R) >droGri2.scaffold_15245 14537789 75 - 18325388 UGAGCAGGGGGGCGCGCAAACGGGCCAAAUGGCUGAAGAAGCGCAUGGUCAGCAAUAUGGUAAGCGGCUUAUAGA---------------- (((((.....(((.((....)).))).....(((((..(......)..))))).............)))))....---------------- ( -18.70, z-score = 0.61, R) >droMoj3.scaffold_6496 10000378 72 - 26866924 UCAGCACUGGGGCCAGAAAAAGAGCCAAAUGGCUGAAGAAGUGCAUGGCCAGCAAAAUGGUUAGCAGCAUAA------------------- (((((.....((((.......).))).....)))))....((((.((((((......))))))...))))..------------------- ( -22.00, z-score = -1.34, R) >consensus UGAGCAGCGGGGCCAGGAAACGGGCCAAGUGGCUGAAGAAGCGCAUGGCCAGCAACAUGGUGAGAGCCAGAAACUUAUAGAA_________ ...........((((........((((....(((.....)))...))))........)))).............................. ( -9.84 = -9.79 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:54 2011