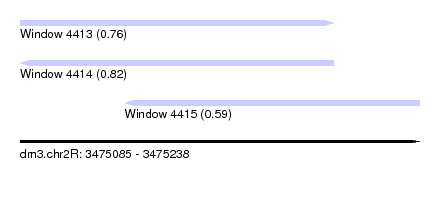
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,475,085 – 3,475,238 |
| Length | 153 |
| Max. P | 0.817148 |

| Location | 3,475,085 – 3,475,205 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Shannon entropy | 0.29783 |
| G+C content | 0.43808 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -21.76 |
| Energy contribution | -23.56 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3475085 120 + 21146708 UUUUGGCAAUGGCAAAACAGGCCGCUACACUACGGUCGGGCUCCCAUAAGUAGAACGACUUAAAGGUGCUCAAUCUGCCAUCAAAUACAUUGUUGCGCACAAUGUACGCUCGAUAGUCGA ..(((((.((((((.....(((((........)))))((((.((..(((((......)))))..)).))))....))))))....((((((((.....)))))))).........))))) ( -37.60, z-score = -2.43, R) >droEre2.scaffold_4929 7518479 120 - 26641161 UUUUGGCAAUGGCAAAACAGGCCGAUCAACUACGGUCGAACUUCCAUAAGUAGAACGUCUUAUAGAUACCUCAUGUGCCAUCAAAUACAUUGUUGUGUACAGUGCACAUUCGAUUGCCGA ..((((((((((((....(((.(((((......)))))....((.(((((........))))).))..)))....)))))).......((((.(((((.....)))))..)))))))))) ( -31.70, z-score = -1.64, R) >droYak2.chr2L 16152888 113 + 22324452 UUUUGGCAAUGGCAAAACAGGCCGAUCAACUACGGUCGAGCUUCCAUAAGAAGAACGUCUUAUGAAUGUUAUAUGUGCCAUUAAAUACAUUGUUGUGUACAGUCGCUAGUCGA------- ..((((((((((((.((((((((((((......))))).)))..(((((((......)))))))..)))).....)))))))..(((((....)))))......)))))....------- ( -32.70, z-score = -2.35, R) >droSec1.super_1 1138131 120 + 14215200 UUUUGGCAAUGGCAAAACAGGCCGCUCCACUACGGUCGGGCUUCCAUAAGUAGAACGACUUAUAGUCACUUAAAGUGCCAUCAAAUACAUUGUUGCACACAAUGUGCACUCGAUAGUCGA ..(((((.((((((.....((..((((.((....)).))))..))((((((......))))))............)))))).......((((.(((((.....)))))..)))).))))) ( -33.80, z-score = -2.01, R) >droSim1.chr2R 2205476 120 + 19596830 UUUUGGCAAUGGCAAAACAGGCCGCUCCACUACGAUCGGGCUUCCAUAAGUAGAACGACUUAUAGAUACUUAAUGUGCCAUCAAAUACAUUGUUGCACACAAUGUGCACUCGAUAGUCGA ..(((((.((((((....((.(((.((......)).))).))((.((((((......)))))).)).........)))))).......((((.(((((.....)))))..)))).))))) ( -30.70, z-score = -1.50, R) >consensus UUUUGGCAAUGGCAAAACAGGCCGCUCCACUACGGUCGGGCUUCCAUAAGUAGAACGACUUAUAGAUACUUAAUGUGCCAUCAAAUACAUUGUUGCGCACAAUGUACACUCGAUAGUCGA ..(((((.((((((.....(((((........))))).....((.((((((......)))))).)).........))))))....((((((((.....)))))))).........))))) (-21.76 = -23.56 + 1.80)

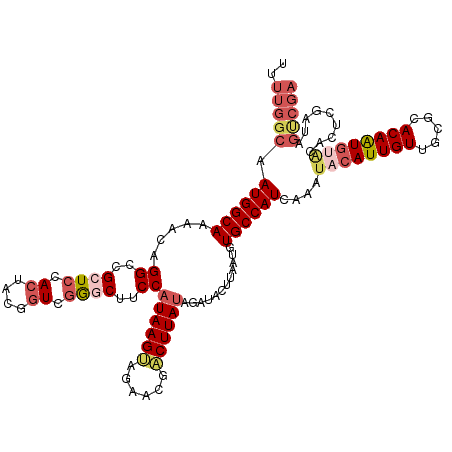
| Location | 3,475,085 – 3,475,205 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Shannon entropy | 0.29783 |
| G+C content | 0.43808 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -25.02 |
| Energy contribution | -27.22 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3475085 120 - 21146708 UCGACUAUCGAGCGUACAUUGUGCGCAACAAUGUAUUUGAUGGCAGAUUGAGCACCUUUAAGUCGUUCUACUUAUGGGAGCCCGACCGUAGUGUAGCGGCCUGUUUUGCCAUUGCCAAAA (((.....)))(((((((((((.....)))))))))..((((((((((((.((.(((.(((((......))))).))).)).)))((((......)))).....)))))))))))..... ( -38.10, z-score = -1.93, R) >droEre2.scaffold_4929 7518479 120 + 26641161 UCGGCAAUCGAAUGUGCACUGUACACAACAAUGUAUUUGAUGGCACAUGAGGUAUCUAUAAGACGUUCUACUUAUGGAAGUUCGACCGUAGUUGAUCGGCCUGUUUUGCCAUUGCCAAAA ..(((((((((..(((((.(((.....))).)))))))))(((((.(..((((.((((((((........)))))))).(.(((((....))))).).))))..).)))))))))).... ( -35.80, z-score = -2.09, R) >droYak2.chr2L 16152888 113 - 22324452 -------UCGACUAGCGACUGUACACAACAAUGUAUUUAAUGGCACAUAUAACAUUCAUAAGACGUUCUUCUUAUGGAAGCUCGACCGUAGUUGAUCGGCCUGUUUUGCCAUUGCCAAAA -------.......((....(((((......)))))..(((((((.....(((((((((((((......))))))))).((.(((.((....)).))))).)))).)))))))))..... ( -25.00, z-score = -0.85, R) >droSec1.super_1 1138131 120 - 14215200 UCGACUAUCGAGUGCACAUUGUGUGCAACAAUGUAUUUGAUGGCACUUUAAGUGACUAUAAGUCGUUCUACUUAUGGAAGCCCGACCGUAGUGGAGCGGCCUGUUUUGCCAUUGCCAAAA ......((((((((((..((((.....))))))))))))))((((......(..(..(((.(((((((((((.((((........))))))))))))))).))).)..)...)))).... ( -38.10, z-score = -2.40, R) >droSim1.chr2R 2205476 120 - 19596830 UCGACUAUCGAGUGCACAUUGUGUGCAACAAUGUAUUUGAUGGCACAUUAAGUAUCUAUAAGUCGUUCUACUUAUGGAAGCCCGAUCGUAGUGGAGCGGCCUGUUUUGCCAUUGCCAAAA ......((((((((((..((((.....))))))))))))))((((......(((...(((.(((((((((((..(((....))).....))))))))))).)))..)))...)))).... ( -35.30, z-score = -1.72, R) >consensus UCGACUAUCGAGUGCACAUUGUGCGCAACAAUGUAUUUGAUGGCACAUUAAGUAUCUAUAAGUCGUUCUACUUAUGGAAGCCCGACCGUAGUGGAGCGGCCUGUUUUGCCAUUGCCAAAA .......((((((..(((((((.....)))))))))))))(((((...................((((((((.((((........))))))))))))(((.......)))..)))))... (-25.02 = -27.22 + 2.20)

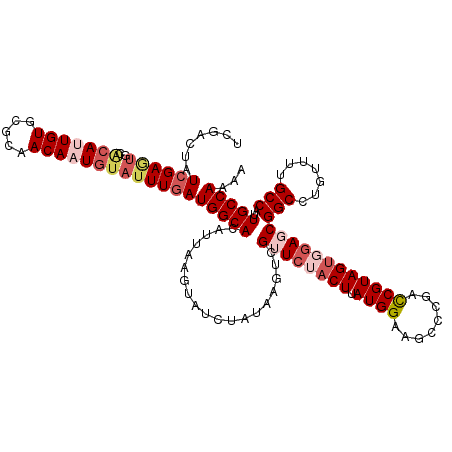
| Location | 3,475,125 – 3,475,238 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Shannon entropy | 0.41243 |
| G+C content | 0.37592 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
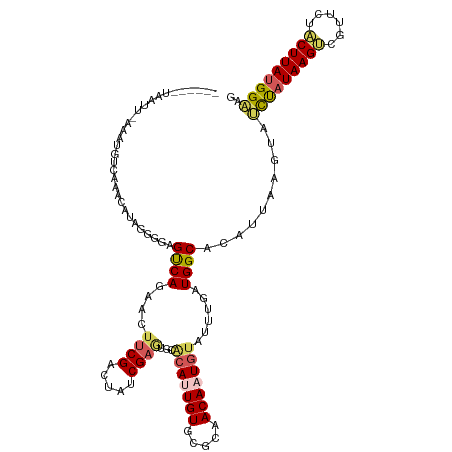
>dm3.chr2R 3475125 113 - 21146708 ------UAAUU-AAAUAUCUGAGAAAGGGGAGCCAGAACUUCGACUAUCGAGCGUACAUUGUGCGCAACAAUGUAUUUGAUGGCAGAUUGAGCACCUUUAAGUCGUUCUACUUAUGGGAG ------.....-....(((((.....((....))..........(((((((..(((((((((.....)))))))))))))))))))))......(((.(((((......))))).))).. ( -28.60, z-score = -1.19, R) >droEre2.scaffold_4929 7518519 117 + 26641161 ---GAAUAUUUCCAAUAUCGAGUAUAGGGGAGCCAGAACUUCGGCAAUCGAAUGUGCACUGUACACAACAAUGUAUUUGAUGGCACAUGAGGUAUCUAUAAGACGUUCUACUUAUGGAAG ---.....(..((.((((...)))).))..)(((........))).(((..((((((...(((((......)))))......))))))..))).((((((((........)))))))).. ( -28.60, z-score = -1.38, R) >droYak2.chr2L 16152928 113 - 22324452 GAUUAAUAUGAUAAAUGUCAAAUAUAGGGGAGUCAGAAUAUCGACUAGCG-------ACUGUACACAACAAUGUAUUUAAUGGCACAUAUAACAUUCAUAAGACGUUCUUCUUAUGGAAG .....(((((.....(((((..(((((...((((........))))....-------.))))).(((....)))......))))))))))....(((((((((......))))))))).. ( -20.30, z-score = -0.82, R) >droSec1.super_1 1138171 113 - 14215200 ------UAAUU-AAAUGUCGAACCUAGGGAUGUCAGAACUUCGACUAUCGAGUGCACAUUGUGUGCAACAAUGUAUUUGAUGGCACUUUAAGUGACUAUAAGUCGUUCUACUUAUGGAAG ------.....-....((((((......(....).....))))))....((((((.(((((.(((((....))))).))))))))))).......((((((((......))))))))... ( -26.90, z-score = -1.02, R) >droSim1.chr2R 2205516 113 - 19596830 ------UAAUU-AAAUGUCAAACAUAGGGUUGUCAGAACUUCGACUAUCGAGUGCACAUUGUGUGCAACAAUGUAUUUGAUGGCACAUUAAGUAUCUAUAAGUCGUUCUACUUAUGGAAG ------.....-.........((.(((.((..((((((((.((.....)))))..(((((((.....))))))).)))))..))...))).)).(((((((((......))))))))).. ( -27.20, z-score = -1.54, R) >consensus ______UAAUU_AAAUGUCAAACAUAGGGGAGUCAGAACUUCGACUAUCGAGUGCACAUUGUGCGCAACAAUGUAUUUGAUGGCACAUUAAGUAUCUAUAAGUCGUUCUACUUAUGGAAG ...............................((((....((((.....))))...(((((((.....)))))))......))))..........(((((((((......))))))))).. (-17.32 = -17.84 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:53 2011